ABOVE: © istock.com, KrulUA
As the acute phase of the COVID-19 pandemic recedes further into the global rearview mirror, life science research—and in particular, the tools that fuel it—continues to forge ahead. The past couple of iterations of The Scientist’s annual Top 10 Innovations featured many products that directly addressed the (hopefully) once-in-a-generation disease outbreak, but also highlighted technological advances that pressed forward even in the face of that massive global disruption.
This year’s winners reaffirm that the research enterprise has not only persevered but gained momentum as the world emerges from the worst that SARS-CoV-2 threw at us. These include technology that can sequence a human genome for $100, highly sensitive imaging platforms for studying individual cells and subcellular compartments, and an assay system that facilitates protein discovery. There are also several tools with a clinical focus, such as personalized sequencing panels to detect residual cancer cells left after tumor removal, a software tracking system for overseeing gene and cell therapies from bench to bedside, and a DNA processing tool that improves technicians’ ability to analyze fetal DNA in a mother’s blood.
We are happy to announce the new products that our panel of independent judges has chosen as this year’s Top 10 Innovations.
NeXT Personal™ Personalis
Small quantities of cancer cells can linger in the body after tumor removal, a phenomenon known as minimal or molecular residual disease (MRD) that, if left undetected, can lead to recurrence. Personalis’s NeXT Personal™ assay, unveiled in December 2021, uses a patient’s own tumors to detect, quantify, and monitor circulating tumor DNA in order to spot MRD and track responses to therapy.
Using 1 mm3 of tumor tissue sampled from a patient, Personalis’s lab performs whole genome sequencing to identify up to 1,800 single-nucleotide variants. These patient-specific mutations are used to design a panel of primers targeting those regions for sequencing in future blood biopsies. The assay also includes primers for other known cancer-related genes, says Dan Norton, associate director for product management at Personalis. “We can see if there are other variants emerging that have a precision therapy associated that may be more effective for that patient.”
Medical oncologist Jonathan Loree of BC Cancer and the University of British Columbia began partnering with Personalis in August to use NeXT Personal in a study of patients who’d had tumors removed from their pancreases or colons, testing the technology’s ability to diagnose disease recurrence “earlier when there is a window of opportunity for patients to still be cured,” he says in an email to The Scientist. Loree says that if the assay could replace conventional diagnostic CT scans and blood tests, “[t]hat has the potential to save money [and] improve outcomes.”
NeXT Personal is currently used in research only, with plans to expand to clinical trial settings next year, Norton says. Personalis declined to provide a cost for NeXT Personal, explaining that the price varies depending on user needs.
KAMDAR: “NeXT Personal offers [the potential to] address a number of tumors that are not fully analyzed by other technologies to help identify and manage a patient’s disease.”

CosMx™ Spatial Molecular Imager NanoString Technologies

The CosMx™ Spatial Molecular Imager (SMI) visualizes and quantifies RNA and protein levels at the single-cell and even subcellular levels. The platform, developed by NanoString Technologies, Inc., allows users to follow a standard protocol to prepare and hybridize specific probes and antibodies to their samples, which can be frozen tissue slices or formalin-fixed, paraffin-embedded slices. In the automated instrument, reporter sets hybridize and are imaged, then the fluorescent dyes are cleaved with UV light and washed off before the next reporter set hybridizes with the sample, allowing researchers to image multiple targets in one sample.
The CosMx SMI, priced at US$295,000, contains a high-resolution microscope and “allows researchers to visualize and quantify 1,000 RNA and 100 protein targets at a subcellular resolution across more than 1 million cells,” Vikram Devgan, senior director of Spatial Genomics Business at NanoString, says in an email to The Scientist. He adds that users can also purchase the AtoMx™ Spatial Informatics Platform, a subscription-based software produced by NanoString, to visualize and analyze the data generated by the CosMx.
“The CosMx is the only instrument that has provided us with the opportunity to simultaneously visualize thousands of genes, at subcellular resolution, and across all cells in a tissue,” says Miranda Orr, an Alzheimer’s disease researcher at Wake Forest University School of Medicine in North Carolina who, after using another NanoString product, bought the CosMx SMI. “We are able to develop maps of the brain at an unprecedented resolution.”
QIAN:“This will transform the in situ spatial biology and molecular pathology fields.”

UG100™ Ultima Genomics

Ultima Genomics announced this May that it had developed technology to sequence an entire human genome for US$100. Thus far, only early-access customers have had the chance to use the company’s new platform, called UG100™, but Ultima Genomics expects to release the product to the broader market in the first half of 2023. Compared to other sequencers, UG100 has several advantages, including higher speed, better efficiency, and less waste, says Josh Lauer, the company’s chief commercial officer.
Lauer attributes many benefits of the UG100 to a unique feature: a circular, open flow cell. Reagents are applied directly to a spinning silicon wafer that distributes them more efficiently than reagents pumped through a traditional flow cell, Lauer explains. In addition, the revolving design increases the speed of data collection and imaging, enabling Ultima’s sequencer to complete one run in about 20 hours, which he says is about twice as fast as existing technologies. “Much like a CD player, this enables ultra-high-speed scanning of genetic material.”
“I'm excited about the throughput of the platform, as well as the cost,” says Reuben Saunders, a genetics graduate student at the Whitehead Institute who collaborated with Ultima Genomics to use its UG100 in recent research. “It’s heralding an exciting era where very large-scale experiments . . . will become accessible methods that can really drive advancements in our understanding of genetics and cell biology.”
Lauer says the $100 per genome cost includes wafers and chemical reagents, but Ultima Genomics declined to release the price of the refrigerator-sized hardware that performs the sequencing.
RAO: “This is the first under $100 genome, and they have achieved it with an innovative use of technology.”

Proteograph™ Product Suite Seer

Proteomic studies have traditionally faced two key challenges, says Rebecca Rutherford, Director of Product Management at biotech company Seer: The need to tag proteins has restricted research to known proteins, and the large, diverse nature of the proteome has made investigating low-abundance proteins difficult. Seer’s Proteograph™ Product Suite, launched in January 2022, addresses both challenges, she says, using a nanoparticle-based assay that allows unbiased sampling of all peptides in a biofluid sample. “The innovation in the Proteograph Product Suite is really around our proprietary engineered nanoparticles that have unique surface functionalization that attract proteins across the entire dynamic range.” This allows researchers to track small molecular changes associated with disease and reveals distinct protein variants produced by post-transcriptional modifications, Rutherford says, enabling the identification of novel and biologically relevant proteoforms.
“From a [discovery] proteomics perspective, the complex liquid biopsies like blood, serum, and plasma that we work with have just been inaccessible,” says Mark Flory of the Cancer Early Detection Advanced Research Center (CEDAR) at Oregon Health & Science University Knight Cancer Institute who collaborated with Seer to test the new platform before becoming the first client to purchase it. Proteograph enables “deep sampling in those very complex liquid biopsy types.” His research team has been applying the platform to a large cohort study of prostate cancer to identify new biomarkers. The product is also being applied to research on lung cancer and Alzheimer’s disease, according to Rutherford.
Seer declined to provide a price for the Proteograph™ Product Suite.
KAMDAR: “Provides access to the proteome in an unbiased way and does for proteomics what next-generation sequencing has provided in genomics.”

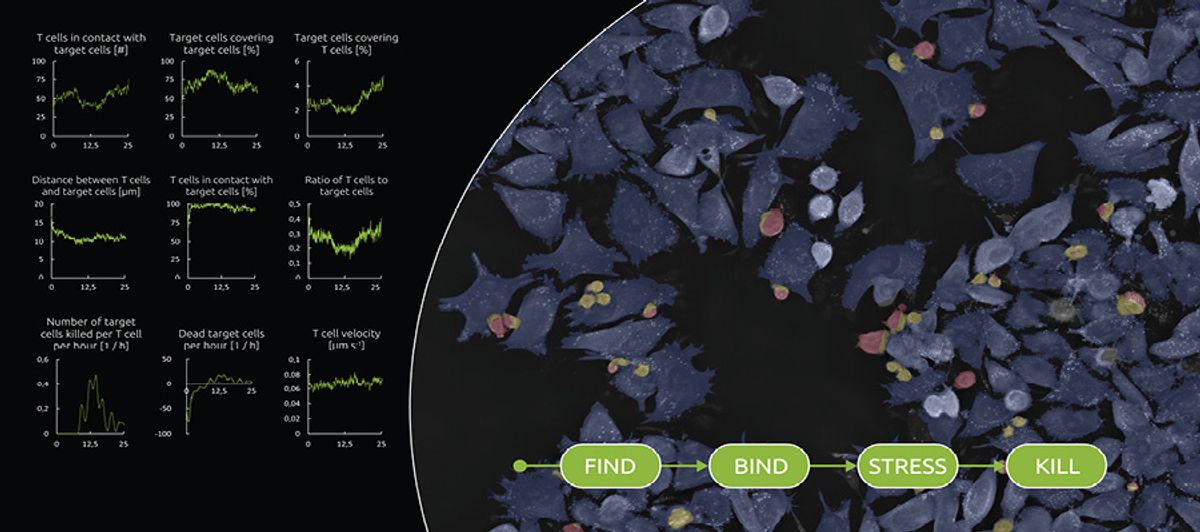
LIVE T Cell Assay Nanolive

Over the past decade, Nanolive has developed imaging platforms based on technologies that reconstruct three-dimensional holo-grams of label-free samples. In September 2021, they launched their first application-specific digital assay, LIVE T Cell Assay, which examines how T cells locate, bind, stress, and kill their targets, such as infected, foreign, or cancerous cells. The assay measures phenotypic and morphological parameters of both the T cells and the target cells, but “what’s really novel about the product is the metrics that we can extract from the interaction between the two,” says Emma Gibbin-Lameira, scientific communications manager at Nanolive. Such information can be very powerful in testing the efficacy of a drug, she adds. For instance, you can assess whether a specific antibody brings T cells closer to the desired target and whether it increases the cells’ killing rate.
Valery Moine, a unit head in the Pharmacology group at Switzerland-based Light Chain Bioscience who collaborates with Nanolive, says he started using LIVE T Cell Assay a year ago to create “marketing videos to highlight and promote the mode of action of bispecific antibodies” developed by his company. More recently, he says he has been using the platform to further characterize these antibodies. The metrics it provides, he adds, are valuable for ranking the best candidates.
Nanolive declined to share the price of the assay, but Lisa Pollaro, the chief marketing officer at the company, writes in an email that “it comes with an annual license with a price in the same range of chemical assay kits available in the market.”
RAO: “A non-end point assay for the rapidly developing field is a huge advance in enabling therapy.”
MARS® Bar Applied Cells, Inc.

One of the most efficient methods to select cells is immunomagnetic separation, where the isolation is based on the presence of magnetic beads attached to specific cell surface antigens. There are several products on the market that apply this method, but a new system presented by Applied Cells, Inc., called MARS® Bar, has various advantages, says Liping Yu, vice president of applications at the company. For instance, MARS Bar works as a closed system with sterile fluidic kits, which makes it “much easier to manage,” says Yu, because its use is no longer restricted to a clean room or biosafety cabinet. Additionally, the device contains three modules, allowing it to process three samples in parallel.
Sergei Rudchenko, an assistant professor at Columbia University who has an ongoing scientific collaboration with Applied Cells, has been using a version of the product, MARS Bar Flex—quite similar but with open fluidics—since February for a protocol that aims to remove naive T cells from peripheral blood mononuclear cells during cell transfusion. According to Rudchenko, who says he is interested in developing the protocol into a clinical application to prevent graft-versus-host disease, MARS Bar Flex achieves about eight times higher recovery of blood cells after depletion of unwanted cells than other published protocols, in addition to “quite competitive purity.”
According to Yu, MARS Bar is customizable but will cost around US$150,000 for the standard three-module configuration once it is available on the market. The company started demoing this new model at customer sites in November, she adds.
RAO: “Very innovative use of magnetic separation technology without fixed magnets allowing a high throughput.”

Single Cellome™ System SS2000 Yokogawa Electric Corporation

The Single Cellome™ System SS2000 by Yokogawa Electric Corporation is an automated subcellular sampling system, which contains both a dual spinning-disk confocal microscope to visualize tissue and a sampling setup to collect whole cells or intracellular components from a single cell.
The system, launched in February 2022 in Japan, the US, and China, can be fully automated, giving users the ability to define which cells should be sampled based on their cytoplasm area, nucleus size, or other morphological features. Cells that fit a particular profile are aspirated and deposited in a 96-well plate for further analysis. The system can also directly sample intracellular components such as organelles or parts of the cytoplasm and combine these insights with whole-cell sampling. The SS2000’s “high-resolution 3D image allows researchers to control the location of [cell and subcellular] sampling in a highly precise way and retains the spatial context,” says Takanobu Kiuchi, head of global marketing at Yokogawa.
“We can sample multiple times from the same cell, you collect a small intracellular sample for metabolomics, and then collect the rest of the cell for single-cell transcriptomics,” says Carla Newman, associate director of Cellular Imaging and Dynamics at GSK, who received an SS2000 from Yokogawa as part of a research collaboration agreement for beta testing, Especially for rare phenotypes, she notes, the ability to target sampling to specific features is highly useful, as well as being able to sample small numbers of patient cells. “It allows for the granularity of the single-cell level to pick up rare events.” Newman adds that the SS2000 is faster and easier than traditional micromanipulators, improving the sampling speed by at least 10 times.
Depending on the technical configuration and required support, the instrument list price ranges from €650,000 (US$651,800) to €725,000 (US$727,088).
QIAN: “This is a highly innovative system coupling high resolution cellular imaging with subcellular sampling technology.”

LightBench® Detect Yourgene Health

The LightBench Detect® is a DNA processing tool that is useful for noninvasive prenatal testing (NIPT), which involves fetal DNA collected from the mother’s blood. To help find the tiny strands of fetal DNA among the clusters of maternal DNA, the LightBench Detect separates the fragments by length, explains Yourgene Health product manager Becky Underwood.
The product employs Yourgene’s imaging system, Ranger Technology, to image the gel and make real-time adjustments to the voltage to optimize strand separation, Underwood says, adding that the LightBench Detect is the only instrument on the market that can use plastic EDTA blood collection tubes, which are cheaper, less susceptible to breakages, and more efficient than the industry standard glass ones. These features yield 50 to 75 percent more fetal DNA per sample, according to Yourgene’s product page.
The instrument, which costs US$50,000, launched in early June and is still in its testing stages with company scientists and a few outside researchers, Yourgene tells The Scientist. Bhavika Patel, director of the Yourgene genomics services lab that is using the product, says its usage of EDTA tubes has significantly cut down the lab’s costs and failure rates, potentially putting less strain on patients and getting quicker results. “It’s quite a nice, neat instrument,” Patel says.
In addition to its use for NIPT, Underwood says the LightBench Detect has a wider range of applications, including scanning blood samples for tumor DNA or infectious pathogens. “We want to positively influence clinical pathways and improve patient outcomes,” Underwood tells The Scientist.
VAN VLIET: “This can be a game changer for widely used diagnostics globally, especially given price point and ease of sample preparation.”

Molecular Cartography™ Resolve Biosciences

Resolve Biosciences’s Molecular Cartography™ workflow is a single-molecule fluorescence in situ hybridization technology that offers a three-dimensional view of gene expression within cells without damaging the tissue section or cell culture sample. The company launched the platform as a mail-in service last year, and that service won a spot in the 2021 Top 10 Innovations list. Resolve Biosciences began installing hardware and software for the fully automated Molecular Cartography workflow in customer laboratories in January 2022, for a cost of US$400,000.
“We’re able to bring the assay into the actual disease state and map interactions at the single molecule level within tissues,” says Jason T. Gammack, cofounder and CEO of the firm. The platform produces high-resolution images of subcellular gene expression, which, in addition to providing unique insights into the transcriptional landscape of the cell, are “quite breathtaking,” Gammack says. “You now see the beautiful symmetry of biology.”
“It is a ready-to-go system that needs very little optimization,” says Jan-Philipp Mallm, head of the Single-Cell Open Lab at the German Cancer Research Center (Deutsches Krebsforschungszentrum, DKFZ). A major advantage of Molecular Cartography is its fully customizable panel of probes, says Mallm, who used the mail-in service before purchasing the full workflow for his laboratory this year. “I think the versatility is a big asset of the system.” Mallm and his colleagues are applying the technology to study the cancer microenvironment, where “a tumor can be regarded as a whole complex tissue and thus needs single-cell spatial resolution in order to understand its function and capabilities.”
KAMDAR: “This view of subcellular gene expression activity can facilitate new insights into the interactions and complexity of critical biological mechanisms.”

OCELLOS 3.0 TrakCel

Cell and gene therapy have started to revolutionize medicine, but they’ve also presented new challenges in tracking the materials involved, including patients’ own cells. To address these challenges, TrakCel introduced a cloud-based software called OCELLOS in February 2021 and released a new iteration, OCELLOS 3.0, this July. “It’s a really simple computer interface that’s easy to access throughout the whole supply chain,” says Matthew Lakelin, a cofounder of TrakCel and the company’s vice president of scientific affairs and product development. OCELLOS 3.0 not only bolsters the safety and quality of such treatments, but it manages the chain of identity and chain of custody data that regulatory agencies require, Lakelin notes.
Edward Armstrong, senior director of quality assurance at Mustang Bio, has been partnering with TrakCel since 2018, using their technology to track Mustang’s autologous CAR T cell products and cell and gene therapies during clinical trials. “Chain of custody and chain of identity are critical to our process and are looked at very heavily by the Food and Drug Administration,” says Armstrong, who is currently switching to OCELLOS 3.0. When Mustang Bio was looking to enroll its first patient, Armstrong considered tracking the products on paper, but quickly realized that “to do on paper what TrakCel does electronically would drive mortal men insane.”
TrakCel’s OCELLOS 3.0 starts at $300,000, but the price may increase based on the amount of involvement and customization desired.
VAN VLIET: “This product can help focus, simplify, and reduce errors in the complex supply chain and task logistics of cell/gene therapy (CGT) development and production.”

THE JUDGES

Kim Kamdar
Managing partner at Medical Excellence Capital, a healthcare-focused venture fund creating and investing in biopharma and diagnostic companies. She began her career as a scientist and pursued drug-discovery research at Novartis/Syngenta for nine years.

Wei-jun Qian
Bioanalytical chemist at Pacific Northwest National Laboratory. His research centers primarily on the development and applications of mass spectrometry–based approaches to better quantify the dynamic changes in protein abundances and protein post-translational modifications in biological and clinical applications.

Mahendra Rao
CEO at Implant Therapeutics. Rao has published more than 200 papers on stem cell research and is the cofounder of the neural stem cell company Q Therapeutics, based in Salt Lake City, Utah. He has served on advisory panels for the US Food and Drug Administration (FDA), as well as for the governments of the US, Singapore, and India on policies pertaining to human embryonic stem cells. He continues to work with the FDA and other regulatory authorities on issues related to pluripotent stem cells.

Krystyn Van Vliet
Michael (1949) and Sonja Koerner Professor in the departments of materials science and engineering and biological engineering at MIT. She also leads the Singapore-MIT Alliance for Research and Technology's Critical Analytics for Manufacturing Personalized-Medicine (CAMP) research team.






