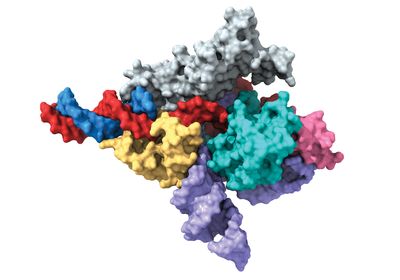
ABOVE: Eukaryotic Fanzor proteins can be used to edit DNA. This image depicts a Fanzor protein in a cryo-EM map where the guiding RNA is purple and the DNA is red and blue. the Zhang lab at MIT, the Broad Institute
Since the bacterial-origin CRISPR-Cas system rose to popularity as a genome editing tool, scientists have wondered whether other genome editors exist in nature. Now, two independent research teams have discovered that Fanzor proteins, which are found across eukaryotic life, function similar to the Cas nucleases in the CRISPR-Cas system, which means that they can be used for genome editing.
“One question that people always ask me is, if CRISPR is so abundant in the microbial prokaryotic world, how come it is nowhere else?” said Feng Zhang, a molecular biologist at Massachusetts Institute of Technology (MIT) and the Broad Institute who led the work published recently in Nature.1 “What’s really exciting here is that Fanzors are in eukaryotes, so there are some systems resembling CRISPR-like systems in eukaryotic cells.”
What’s really exciting here is that Fanzors are in eukaryotes, so there are some systems resembling CRISPR-like systems in eukaryotic cells.
—Feng Zhang, Massachusetts Institute of Technology
Scientists have known about Fanzors’ presence in eukaryotic life for a while,2 but they didn’t know their evolutionary origins or how they worked. They suspected, however, that Fanzors evolved from the same group of proteins as the Cas enzymes, a class called obligate mobile element-guided activity (OMEGA). Consequently, both Zhang and his former colleagues Omar Abudayyeh and Jonathan Gootenberg at MIT, who published similar work in a preprint a few weeks before Zhang published,3 began by searching sequence databases for Fanzor-like proteins to establish a phylogenetic tree.
The OMEGA suspicion held up; the teams found that bacteria likely passed OMEGA proteins to eukaryotic organisms via viruses and that these proteins then evolved into Fanzors. “That would make them almost cousins to the CRISPR systems,” said Abudayyeh. Fanzors were also structurally similar to both OMEGA and Cas proteins. Together, these findings hinted that Fanzors could have the RNA-guided endonuclease activity that enables genome editing.
Both teams put their hypotheses to the test. Zhang’s group tested Fanzors from soil fungi, algae, unicellular eukaryotes, and a clam, whereas Abudayyeh and Gootenberg focused on one from a giant virus. All of the proteins exhibited RNA-directed DNA cleavage in vitro, and although only some worked in human cells, this proved that Fanzors have genome editing abilities. Zhang’s team further engineered one of them to improve its editing efficiency.
Zhang, who also led the first demonstration of CRISPR in eukaryotic cells,4 said that the most interesting part was not how Fanzors are like CRISPR, but how they are different. “Differences are beautiful and powerful because there are a lot of different things we want to do,” he said. Gootenberg agreed. “Because [Fanzors] come from a eukaryotic system, there’s the thought that they could potentially have some advantages, like having better function in eukaryotic cells,” he said.
It is too early to say where Fanzors might offer advantages or limitations compared to existing systems, but there are hints. For example, Fanzors are smaller, which might make delivery of genome editing therapies easier, Zhang pointed out. At the same time, their similarity to Cas12 might make them trickier to turn into nickases used in base and prime editing,5 which are usually based on Cas9.
This would almost make them cousins to CRISPR.
—Omar Abudayyeh, Massachusetts Institute of Technology
Answering those questions won’t only have implications for applications in human cells. Geneticist Rodolphe Barrangou from North Caroline State University who was not involved with the work thinks that the discovery could have big repercussions for editing other eukaryotic systems too. “I love that it’s widespread in eukaryotes because it presumes that it’s compatible with all kinds of eukaryotic environments,” he said.
The fact that Fanzors appear prevalent in eukaryotic life implies that Fanzor systems could be suited for applications in eukaryotes beyond gene therapy, such as engineering plants for more sustainable agriculture. “I am equally, if not more intrigued in some ways about how we can employ these genome editors outside of humans,” Barrangou said.
In addition to its applications, there is lots of Fanzor biology and evolution still to understand. “To figure out their biological role is the obvious next step,” said Abudayyeh. CRISPR is involved in the bacterial immune system, but so far, it looks like Fanzors might play a different role related to maintaining jumping genes in eukaryotes.
As scientists continue sequencing the genomes of organisms, they may find even more different molecular systems and capabilities to discover. Zhang, Abudayyeh, and Gootenberg all intend to keep looking for more as-of-yet undiscovered cousins to CRISPR and Fanzor. “We are very interested in understanding these differences and trying to find new processes to study and maybe engineer to do something useful,” Zhang said. “In a way, it’s like treasure hunting out of all the things that nature has created.”
References
- Saito M, et al. Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature. 2023;1-3.
- Bao W, Jurka J. Homologues of bacterial TnpB_IS605 are widespread in diverse eukaryotic transposable elements. Mob DNA. 2013;4(1):1-16.
- Jiang K, et al. Programmable RNA-guided endonucleases are widespread in eukaryotes and their viruses. bioRxiv. 2023.
- Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819-823.
- Koonin EV, et al. Discovery of Diverse CRISPR-Cas Systems and Expansion of the Genome Engineering Toolbox. Biochemistry. 2023.