ABOVE: modified from © ISTOCK.COM, Design Cells
When the Human Genome Project published the first draft of the human genome sequence in 2001, many researchers expected to be able to pinpoint protein alterations that would explain the distinctive features of human brains compared to those of other animals—larger size relative to the body, increased neuronal connectivity, and other contributors to our superior cognitive complexity. Instead, “it was frustrating to see how few protein-coding genes exist,” says Geraldine Zimmer-Bensch, a neuroepigeneticist at Rheinisch-Westfälische Technische Hochschule Aachen in Germany, “and even more frustrating, how little difference there is between the mouse and the human protein-coding genome.” Yes, there are proteins and variants of proteins that are unique to our species, she says, but there simply aren’t enough of them to explain humans’ singular cognitive prowess.
This was particularly surprising because at least a tenth of the human proteome consists of proteins whose main function is in the brain—some estimates say it’s more like a third.
According to Zimmer-Bensch and an increasing number of neuroscientists, the missing piece of the puzzle is RNA—specifically, the myriad RNAs that don’t code for proteins, such as long noncoding RNAs (lncRNAs) and microRNAs (miRNAs). Noncoding RNAs are likely protagonists in our brain’s evolutionary story because they are pivotal regulators of gene expression, especially during development, experts say. Changes in traits such as tissue size and shape are easily made by tweaking when and in what cells different proteins are made—precise alterations that generally occur as the result of changes in noncoding regions of the genome, researchers are finding.
RNA transactions are critical to brain function.
—John Mattick, University of New South Wales
And RNAs aren’t just stars of the evolutionary and developmental past; they are essential for brain functioning now, and evidence is mounting that regulating gene expression is just one of noncoding RNAs’ many neurological tasks. For instance, some noncoding RNAs are actively transported to the ends of axons to play roles completely divorced from gene expression. Plus, notes Zimmer-Bensch, noncoding RNAs can be passed from cell to cell via vesicles and junctions. “The functional diversity [of noncoding RNAs] is tremendous and impressive.”
Research into how RNAs function in the brain has progressed more slowly than the study of protein function, however. For one thing, RNA is less stable than both DNA and the proteins they encode, and many RNAs are only expressed at low levels in specific tissues or cells, making them difficult to detect. “They’re super selective about when and where they’re expressed,” says Timothy Bredy, a molecular neuroscientist at the University of Queensland in Australia. Moreover, scientists don’t yet have a complete grasp of the total number of RNAs encoded in the genome, and novel RNA forms continue to be discovered. But now, cutting-edge sequencing technologies are giving researchers unprecedented insights into cells, allowing RNA studies to be conducted on the spatial and temporal scales needed for the discipline to begin to catch up to protein biology. And findings from this work are pointing to an inevitable conclusion: RNAs rule the brain.
Complex Brains Echoed in RNABursts in microRNA (miRNA) diversity often line up with sudden increases in morphological complexity, especially in the context of the nervous system. In a 2022 bioRxiv preprint, researchers uncovered an miRNA repertoire expansion (orange) in the ancestor of coleoid cephalopods—the group that includes squids and octopuses, generally thought to be more intelligent than any other invertebrates—on par with ones seen in the ancestors of vertebrates (blue) and placental mammals (green). 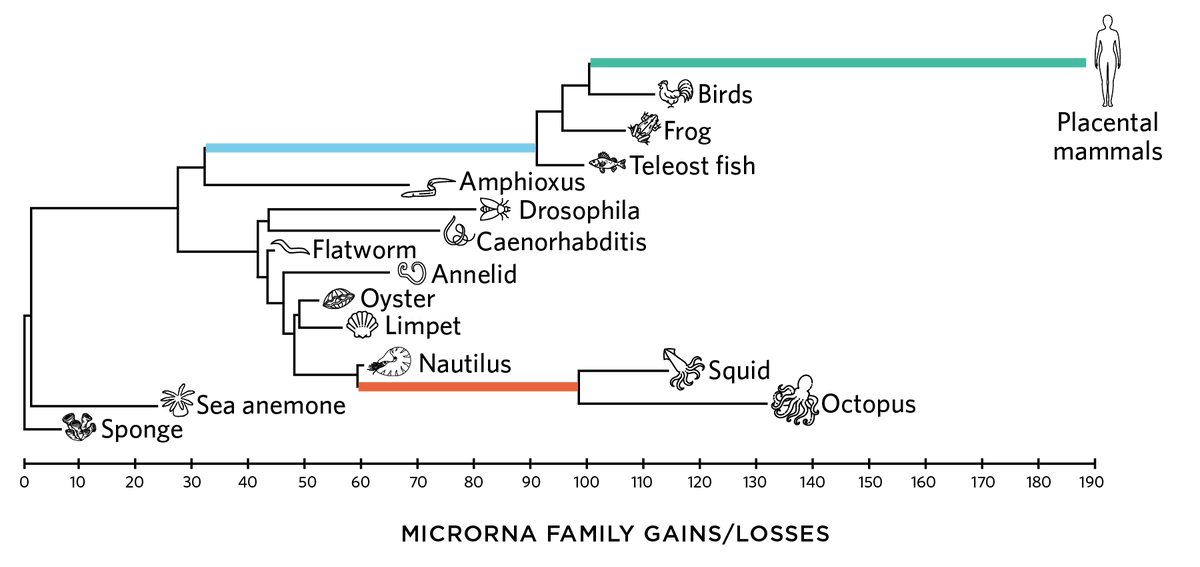 the scientist staff; Icons: © ISTOCK.COM, MaskaRad, GreenTana, Nadiinko, vectorforjoy, Hachio Nora, NatuskaDPI |
Noncoding RNAs as master regulators in brain
According to Debra Silver, a developmental neurobiologist at the Duke University School of Medicine in North Carolina, researchers have for nearly half a century been exploring the idea that regions of the genome that don’t encode proteins may play an outsize role in the brain. But it wasn’t until more-recent technological advances, especially in RNA sequencing and single-cell analyses, that scientists could start to study noncoding RNAs in more detail. This research has led to a shift in understanding of how the brain evolved, Silver says. “In the last 10 years, the idea that RNAs can have an impact, and that layers of regulation between a DNA and a protein are meaningful, has gotten a lot more attention.”
Dartmouth College paleontologist Kevin Peterson and his colleagues have been studying miRNAs phylogenetically, looking for how changes in miRNA inventories map to evolutionary transitions. In addition to finding that miRNA repertoires tend to increase in the genomes of different animal groups over evolutionary time, the team discovered that “there are certain places in evolution where you just had inordinate numbers [of miRNAs] added to a genome,” Peterson says. “And these just happened to coincide with places on the tree where you get these big, obvious jumps in complexity.” This includes a burst of 179 miRNA genes that appeared in the primate lineage after it split from mice.
“Complexity, in general, is kind of defined by an increase in cell types,” Peterson explains, and an increase in miRNAs could support the rise of additional cell types by sculpting novel cellular moieties from the same pool of molecular parts.
Peterson and his colleagues recently found a similar trend among octopuses and their soft-bodied kin, collectively referred to as coleoid cephalopods. In a 2022 bioRxiv preprint, the team reported finding at least 89 miRNA families in cephalopod genomes that were not present in their shelled nautilus cousins, bringing the total number of miRNA families to at least 135. That’s more than usual for an invertebrate and on par with miRNA numbers seen in fish, amphibians, and birds. Furthermore, RNA sequencing from different tissues within cephalopods revealed that most of the novel miRNAs are expressed in the animals’ brains, strongly suggesting they help steer the organ’s development and functioning—all of which is very reminiscent of what’s known about other miRNA expansions linked to neurological and cognitive complexity in distant taxonomic groups.
University of California (UC), Santa Barbara, neuronal cell biologist Kenneth Kosik, who was not involved in the work, says he thinks “very highly of the paper,” and that “it’s really very exciting to see these phylogenetic rapid expansions of microRNAs and microRNA families. It’s really just intrinsically interesting.” In a 2018 review on miRNAs and brain development, Kosik and UC San Francisco collaborator Tomasz Nowakowski refer to miRNAs as “an evolutionary cauldron.” Silver, who also did not participate in the research, agreed. “There’s [a] way of thinking that nature reuses strategies that work over and over, and this could be a nice example of that.”
Peterson says he’ll “go out on a limb” to say that miRNA expansion may be a prerequisite for physiological complexity, including the neurological intricacy that characterizes human brains. It’s not clear that miRNAs are special in this regard, though, notes John Mattick, an RNA biologist at the University of New South Wales in Australia. Rather, the expansion and alteration of all kinds of noncoding RNAs were likely essential to brain evolution, he says. “The more complex the species, the bigger the junk DNA or noncoding RNA repertoire is, so that was how evolution climbed the mountain of developmental complexity,” he speculates, adding that “my expectation is they’re all important, that they’re part of a regulatory fabric.”
The Multitudes of Noncoding RNAThe term “noncoding RNA” is a catch-all for sequences in the genome that are transcribed but typically not translated. These molecules, which account for the majority of the transcribed sequences in the genome, are now thought to play key roles in brain evolution and function. Noncoding RNAs can be classified based on their size, structure, location, or function, with dozens of different kinds described to date. Here are four types of noncoding RNA frequently studied in brain tissues. | |
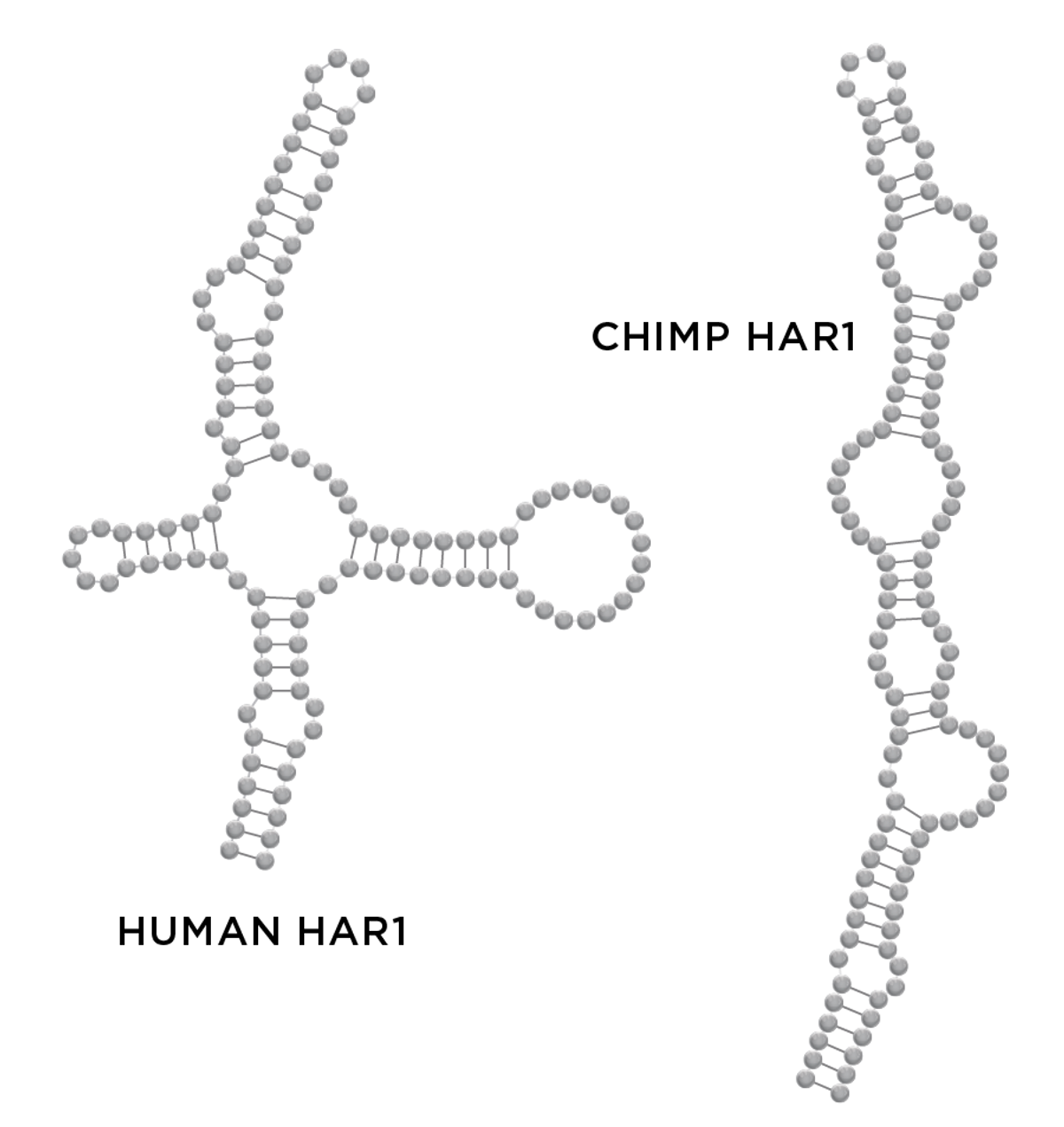 | Long noncoding RNAsLong noncoding RNAs (lncRNAs) are generally described as any noncoding RNAs greater than 200 nucleotides in length. Because of their variable size and composition, they can have complex shapes and perform a variety of cellular activities, though most lncRNAs await functional investigation. Example: The human and chimpanzee versions of a lncRNA called HAR1 differ by 18 nucleotides, which impacts the molecule’s secondary structure. The human version is predicted to be more stable, but exactly how that translates into differences in brain form or function isn’t yet clear. |
 | MicroRNAsMicroRNAs (miRNAs) are small noncoding RNAs of just ~20–26 nucleotides (teal) that are cleaved from larger precursors. Their most well-described function is the regulation of gene expression via binding to messenger RNAs, where they generally inhibit translation and, therefore, reduce the amount of protein produced from a given gene. Example: Overexpression of miRNA-124 leads to Alzheimer’s-like pathologies in mice, and elevated levels of the miRNA are found in the brains of people who died from the disease. |
 | Circular RNAsAs the name suggests, circular RNAs (circRNAs) are noncoding RNAs with joined ends, creating a more stable, circular molecule. Many questions remain as to the functions of circRNAs, but some are known to bind miRNAs, likely acting as sponges to modulate the miRNAs’ translation-suppressing effects. Example: The circRNA CDR1-AS fine tunes neuronal development in humans, binding microRNAs (teal) highly expressed in secretory neurons that regulate developmental gene expression. |
 | Transfer RNAsTransfer RNAs’ primary job is to shuttle amino acids to growing peptide chains during translation. In the brain specifically, there’s emerging evidence that modifications to tRNAs play important roles in neuronal health and disease. Furthermore, tRNA fragments—small chunks from tRNA breakdown—seem to have their own functions, including in neurodegeneration. Example: When researchers exposed Drosophila neuron cultures to synthetic tRFGln-CTG (teal)—a fragment of the tRNA for glutamine—the cells swelled and died, suggesting the fragment could play a role in neuronal necrosis. |
Noncoding RNAs: Neurological jacks-of-all-trades
In most cases, researchers are still trying to understand the function of particular RNAs that appear to have evolutionary significance. When it comes to the evolution of human brains specifically, many of the relevant RNAs fall into what are known as human accelerated regions (HARs): stretches of the genome with mutation rates that increased significantly after humans split from chimpanzees. Take HAR1, for example. It’s a 118-base-pair-long RNA that is expressed in certain neurons during key stages of the development of our uniquely complex cerebral cortex. It contains 18 substitutions that differentiate the human and chimp versions of HAR1 and that scientists have discovered lead to dramatically different molecular structures, which in turn affect the RNAs’ stability and, probably, function. But exactly what HAR1 does and what proteins or RNAs it interacts with remain unclear.
A common hypothesis for how these RNAs function is in the control of gene expression, especially during brain development. Individual noncoding RNAs can alter the expression of multiple genes, meaning that small changes in the RNAs can have cascading effects. For example, miRNAs, short (20–26 base pairs) molecules whose most well-studied function is to bind to messenger RNAs (mRNAs) and interfere with their translation into proteins, may each have hundreds of targets—which means even single base changes could impact the expression levels of as many genes.
“It’s sort of like hitting, for lack of a better term, almost a master regulator of gene expression,” says Silver. “And by doing that, it’s going to influence gene expression of its targets likely in a very cell-specific, tissue-specific, timing-specific fashion, and that itself could then affect expression of downstream targets below that.” Because of this, she adds “even though our genomes of human and, say, chimpanzees are remarkably similar globally at the DNA level, there is a whole host of regulatory changes at the RNA level that are likely to contribute synergistically to human-specific traits.”
Kosik’s work on miRNAs has found that miRNAs may sculpt brain size and shape—and therefore, cognitive complexity—via alterations to the cell cycle. “If you’re controlling the cell cycle, then in some ways you also are controlling cell divisions, and the number of neurons that are being made,” he explains. “And we know that in primates, the number of neurons increased a lot. Same is true in cephalopods.” In addition to cell number, humans are unique in the number of different neuronal cell types and other important cells in the brain, Kosik adds, and different cell types tend to have distinct miRNA profiles. “MicroRNAs are important in development and taking precursor cells along the path to various terminal differentiated outcomes . . . so they do have some correlation with cell specialization in the brain.”
Of course, noncoding RNAs aren’t solely expressed during brain development. In addition to steering neurogenesis, “RNA transactions are critical to brain function,” says Mattick. The majority of genetic loci associated with variations in neuropsychiatric function produce noncoding RNAs, he points out. And specific RNAs have been proven to play pivotal roles in activities like memory formation, and especially, in creating the high degree of neuronal plasticity that is a hallmark of human brains. “The brain is rewiring itself on the fly in response to experience,” Mattick explains, by tweaking the RNA regulation systems that acted earlier in life to steer development.
Sathyanarayanan Puthanveettil, a molecular neuroscientist at the University of Florida Scripps Biomedical Research, agrees. “The role of the transcription and translation is very, very vital, not only for wiring the brain, but also for remodeling the brain,” he says. Because noncoding RNAs regulate those processes during development, he adds, it makes sense that they’re easily repurposed to regulate plasticity throughout the life of the brain.
For instance, Bredy and colleagues found in 2022 that neurons activated by fear-related learning in the mouse prefrontal cortex express a particular lncRNA, one that is encoded within an enhancer for the gene Nr4a2, which is directly involved in memory. Knocking down this lncRNA in mice didn’t affect the initial fear-related learning, but did impair fear extinction in response to behavioral training aimed at reducing the previously conditioned fear response. Further study of this RNA, which the team dubbed ADRAM (activity-dependent lncRNA associated with memory), revealed that it coordinates the activity of Nr4a2 by guiding and scaffolding proteins that epigenetically activate the gene.
In the last 10 years, the idea that RNAs can have an impact . . . has gotten a lot more attention.
—Debra Silver, Duke University School of Medicine
Furthermore, while regulation of gene expression is arguably the central function of noncoding RNAs, experts are quick to note that the scientific understanding of RNA biology is in its infancy, and there are likely many other important things that RNAs do in the brain. Puthanveettil and his colleagues discovered that the lncRNA Gm38257 does something rather unexpected: Instead of simply regulating gene expression, it binds to proteins involved in structuring synapses. These RNA-protein complexes are actively transported by the cell’s cytoskeleton to the tips of hippocampal neurons, where the proteins alter dendritic structure. Because of this, the team renamed the RNA ADEPTR (for activity-dependent transported lncRNA), and when the team knocked it down experimentally in mice, those animals’ neuronal plasticity in response to activity was reduced.
Bredy points out that there are now numerous examples of noncoding RNAs, especially lncRNAs, that are similarly transported to the tips of neurons. And “they’re doing wild stuff out there,” he says, including coordinating the trafficking and clustering of RNA granules, which act as reservoirs for RNAs that can reshape local translation in response to experience, his group showed.
Bredy notes that in this example, the lncRNA, called Gas5, was alternatively spliced and therefore had a different form and function at the synapse than in the nucleus, adding that it’s not uncommon for noncoding RNAs to be edited in this way, just as messenger RNAs are. “Another layer of biology that we’re only just starting to come to realize is that these RNAs . . . assume massively complex structures, which then dictate how they’re functioning in space and time,” Bredy says.
He adds that these RNAs are not fixed entities; they can be edited or altered in biologically meaningful ways and have different activities over time. “We need to be thinking [about] the temporal dimension now, because they can be doing things that are functionally discrete depending on the stage of their lifecycle.” He notes, for instance, that some lncRNAs can join ends and become circRNAs, one of the most recently discovered and poorly understood kinds of noncoding RNA. When he and his colleagues profiled such circular RNAs that were induced synaptically in response to learning in mice, they uncovered one that “if you overexpress it, you get a massive enhancement in memory. So we’re looking at it as a lead candidate for a memory enhancer,” he says, noting that these observations are not yet published.
Even tiny chunks of larger RNAs once thought to be junk may be important, Puthanveettil notes. For instance, he says, researchers once thought that tRNA fragments were just waste from the breakdown of tRNAs. However, as researchers began experimenting with the fragments in cell cultures and in animal models, they discovered that “they do actually produce a physiological change,” he says. “They actually function almost like a protein.”
Yet another potential function for noncoding RNAs, Zimmer-Bensch adds, is in cell-to-cell communication, noting that these molecules are often found in extracellular vesicles, which are known to participate in intercellular signaling. While there has not been much research into this phenomenon to date, studies are finding distinct noncoding RNA profiles in the extracellular vesicles emitted by brain tumors, suggesting they could play a role in tumor progression by modulating the activities of surrounding cells. “What I think is really important and still underinvestigated is their potential for making communication between cells,” says Zimmer-Bensch.
Not So NoncodingNoncoding RNA may be a bit of a misnomer. At least some lncRNAs, circRNAs, and transcripts of other so-called noncoding genomic regions do, in fact, contain open reading frames that code for micropeptides. The coding-noncoding nomenclature for RNAs arose in the early days of genomic sequencing. “That’s kind of human nature there, to have to compartmentalize everything,” says University of Queensland molecular neuroscientist Timothy Bredy. But when it comes to the diversity of forms RNAs can take, researchers now know that such restrictive boxes just don’t capture reality. “We have to come up with a new way to describe them—like multidimensional, or multifunctional RNA species,” he says. Micropeptides don’t appear to be mere translational accidents or otherwise useless strings of amino acids. Experiments in mice have found that micropeptides are produced in the central nervous system and that some are enriched in behavior-related brain regions, sometimes with marked impacts on cells. Recent work suggests that noncoding RNA–derived micropeptides can influence everything from cell proliferation to autoimmune inflammation. While researchers have only begun to characterize the potential roles they play in health and disease, it seems likely that these not so noncoding genes have neurological importance. |
Noncoding RNAs may play roles in brain disease
The growing appreciation of noncoding RNAs’ participation in essential brain functions has led some researchers to suggest that they are likely underappreciated contributors to neurological disease, Mattick says. “Nearly all the clinical trials on the proteins that have been fingered in Parkinson’s, or Alzheimer’s, et cetera, have failed. I think they’re looking in the wrong place. [Proteins are] involved, but it’s really aberrant RNA metabolism that’s causing these problems in the brain—not surprisingly, because the brain is running on RNA controls.”
Silver points out that there’s “a lot of good evidence for micro-RNAs and lncRNAs being linked to, particularly, nerve degeneration.” For instance, researchers examining the brains of a mouse model of Alzheimer’s disease found that the levels of miR-124 are 3.5 times higher than normal in hippocampal cells. They found similarly increased expression in postmortem brain tissues from people who had Alzheimer’s, and when they overexpressed miR-124 in non-Alzheimer’s mice, the animals exhibited learning difficulties, poor memory retention, and a reduction in dendritic spine density—all of which are considered Alzheimer’s-like pathologies. Meanwhile, knocking down miR-124 in the mice reduced their symptoms. Other miRNAs and lncRNAs have been linked to Alzheimer’s, Parkinson’s, and other neurodegenerative diseases.
Noncoding RNAs have also been tied to psychiatric disorders. Perhaps the most well-characterized is the lncRNA Gomafu (also known as RNCR2 or MIAT). Studies have found it is involved in eye and brain development, but it’s also been directly tied to anxiety in mice. “If you knock it out, the animals show exaggerated phenotypes associated with neuropsychiatric disease,” explains Bredy. Indeed, Gomafu is involved in alternative mRNA splicing, and mutations or low expression of the RNA can lead to the aberrant splicing of transcripts seen in schizophrenia. Because of this, researchers have even suggested Gomafu could serve as a potential biomarker for the condition.
Yet other noncoding RNAs have been linked to sundry neurological conditions, including learning disorders and autism spectrum disorder. Most lack mechanistic explanations for their contributions to disease, but that’s just because the field is “at the very edge [of that] right now,” Bredy says, adding that lots of findings “will be coming out within a year. There’ll be a pile of papers on it.”
Silver agrees. “I think we’re still at the cataloging stage.” While sequencing technologies and bioinformatic techniques have finally made it possible to detect small amounts of RNA in cells, she adds, there is so much work to be done to study noncoding RNAs to the level of detail that exists for proteins. The “most interesting but also the most challenging” step is to determine the functions of the thousands of noncoding RNAs that have yet to be explored.
One of the trickier aspects of studying noncoding RNAs is that they don’t act alone. Rather, they function in networks and systems in cells that can be tricky to recapitulate in experimental models. Probing the function of a primate- or human-specific noncoding RNA by expressing it in a mouse model “can be really telling,” says Silver, “but it’s not the same as removing it from the primate.”
Puthanveettil also comments on the limitations of studying noncoding RNAs in animal models. What’s required, he says, is a “very systematic, thorough analysis” of these molecules using the diversity of tools now available. Fortunately, that kind of work is now accelerating, he adds. “I would say, the way things are going, in another 10 years, there’ll be huge advances.”