ABOVE: modified from © istock.com, DrAfter123, MicroStockHub
He has the most studied genome of any person, but many researchers have never even considered the fact that he exists. His identity is unknown, and he is largely unremarkable, biologically speaking. Instead, his significance to genomics stems from a single act. In March of 1997, he responded to an advertisement in The Buffalo News that promised a “tremendous impact on the future progress of medical science.” He subsequently donated a blood sample from which scientists pieced together the majority of the human reference genome compiled by the Human Genome Project (HGP), with the rest supplied by a small number of additional donors.
The advertisement’s promise was no hyperbole. Today, the reference genome occupies a central position in biomedical research. It provides the common language that scientists and clinicians use to synthesize findings from different genomics studies. For many genomics researchers, its ubiquity has lent it a sense of finality and completeness. This monumental technical achievement has morphed into a piece of basic scientific infrastructure—depended on completely but rarely considered explicitly.
Despite these successes, the human reference genome’s shortcomings have gradually become evident over time. These deficiencies come in multiple forms. Some are issues of accuracy: Parts of the reference do not necessarily reflect any one individual’s genome correctly. Some involve diversity: Because the current reference genome is based on only a handful of individuals, it correctly represents some individuals’ genomes but not others’, which risks biasing genetics-based research. Paradoxically, the human reference genome’s strengths also multiply these weaknesses. Its ubiquity in research means that the biases induced by its shortcomings pervade the entire field of human genomics.
The ubiquity of the human reference genome in research means that the biases induced by its shortcomings pervade the entire field of human genomics.
After many years of discussion among human geneticists and computational biologists, this community is now attempting to conclusively address these problems. Two large-scale, international collaborations are at the forefront of these efforts: the Telomere-to-Telomere Consortium and the Human Pangenome Reference Consortium, in which one of us (J.E.) is a member. In addition to fleshing out researchers’ understanding of the human genome, these projects are rewriting the book on some of the most fundamental research methods in genomics.
How the Human Genome Project started
Although pieces of the human genome were sequenced by 1990, when the HGP was formally launched, the project was the first large-scale attempt to identify the sequence of the vast majority of the specific 3-billion-nucleotide-long series of As, Cs, Gs, and Ts that make up a human genome. Many colloquially refer to this effort as “sequencing the human genome.” However, this process actually involves two steps: the sequencing of DNA, in which a specialized machine reads the sequences of DNA fragments, and assembly, when researchers piece together these short sequences to construct larger contiguous sequences (contigs, for short) and eventually a full genome by looking for overlap between two reads—much like putting together pieces of a jigsaw puzzle. (See illustration.) Both steps are challenging in their own ways. Sequencing usually involves complex chemical reactions that, unless cleverly engineered, can be prohibitively time-intensive. Assembly requires algorithms that can process millions of sequences efficiently.
Over the span of a decade, researchers working on the HGP painstakingly sequenced and assembled the human genome, primarily using bacterial artificial chromosomes (BACs): human sequences on the order of 100 kilobases that were transferred into bacteria and then amplified by cultivating the bacterial colony. The process of breaking up the human genome into BACs was laborious, but it reduced the genome assembly problem to a manageable scale—closer to the size of viral and bacterial genomes that had been successfully assembled previously using traditional Sanger sequencing. (See illustration.)
In 2001, the HGP published a draft human genome sequence to considerable acclaim and excitement. However, as the name implied, this genome was far from a finished product. Firstly, the draft only targeted the genome’s euchromatic sequences: DNA that is stored within the nucleus in a loosely packed way. Conversely, heterochromatic sequences, which are tightly packed at the periphery of the nucleus, are characterized by extreme levels of repetition, and they were deemed to be too difficult to assemble. Following the puzzle metaphor, imagine trying to complete a large region of solid blue sky. Another limitation of the HGP’s initial draft genome was that it covered only 94 percent of euchromatic sequences; the genome sequence was interspersed with 150,000 gaps where the sequence could not be determined. In some cases, the precise ordering or orientation of the assembled sequences was also unresolved.
Over the next few years, the HGP engaged in an intensive “finishing” effort to fill in these gaps largely using the same technology and assembled a full 99 percent of the euchromatic sequence. In the end, the number of gaps was reduced from 150,000 to 341, primarily around regions with large-scale duplications and the challenging heterochromatic sequences of the centromeres and the subtelomeres, the sequences at the ends of each chromosome just before the telomeres themselves.
In 2007, the Genome Reference Consortium (GRC) was formed to assume stewardship of the human reference genome. It oversaw continued gradual improvements as new technologies and approaches, such as sequencing by synthesis and long reads, highlighted previous errors and filled in missing sequences.
Assembly Versus AlignmentAfter sequencing fragments of DNA to obtain reads, most genomic pipelines follow one of two steps. The reads can be de novo assembled to construct longer stretches called contigs from scratch, with overlapping sequences on the ends dictating which read pieces belong next to each other (left). Alternatively, reads can also be aligned to a reference genome to identify small genetic variations (right). Where de novo assembly can be thought of as assembling a puzzle without the use of the picture on the box, alignment is the equivalent of piecing together a puzzle by looking at that picture. However, because a singular reference genome fails to capture all of the genetic diversity across humans, some sections of DNA might not be able to align to the reference genome well. 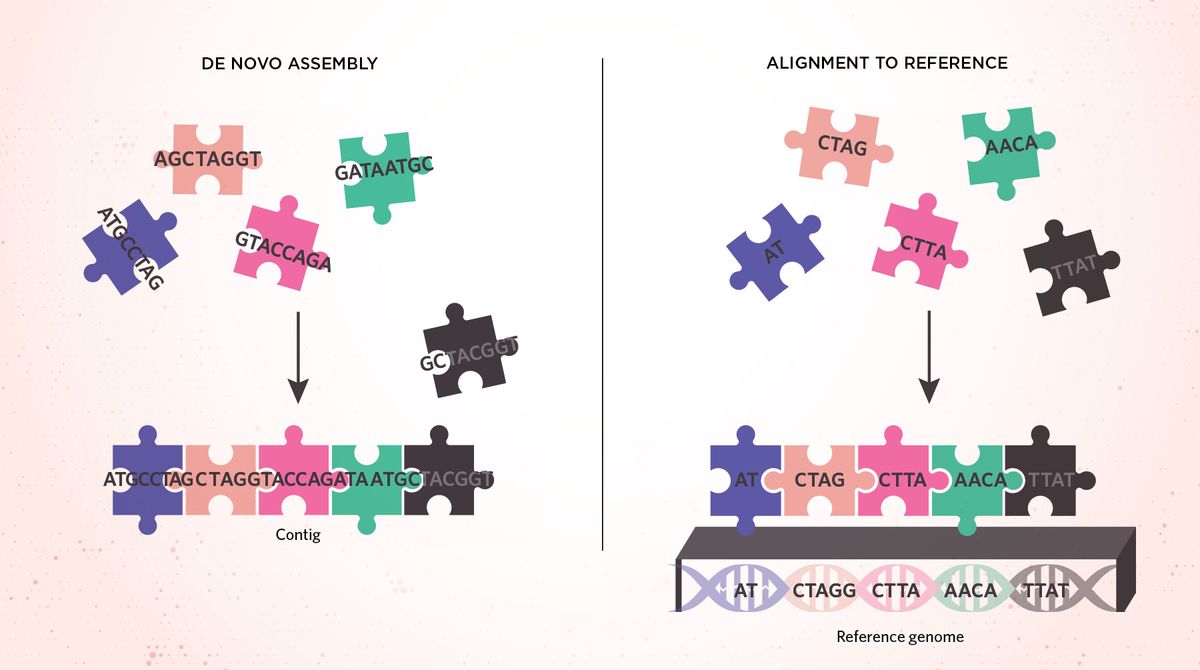 modified from © istock.com, filo |
These earlier efforts were a boon to genomic research. For one thing, they enabled the most widely used modern method for studying human genetics of disease on a population scale: the genome-wide association study, or GWAS. The goal of a GWAS is often to compare the genotypes of a diseased “case” cohort, and a healthy “control” cohort to ascertain genetic contributors of a certain disease. To draw that comparison, genomicists identify the region on the reference genome that is the most similar to each read’s sequence and presumably the region from which it originates. This process, called alignment, is much like putting together a puzzle with the help of the picture on the box. Researchers then look for differences between each sampled genome and the reference and compare the differences identified in the case and control cohorts. Over the past 10 years, GWAS has discovered genetic contributors to hundreds of phenotypes, including psychiatric conditions such as autism and schizophrenia, and diseases that many people previously believed to be entirely environmental such as obesity. GWAS has even helped unearth the genetic roots of human characteristics as complex as being a “morning person.”
But the current human reference genome, HG38, remains incomplete despite the GRC’s efforts to complete it. Some of the 341 remaining gaps proved stubbornly intransigent, especially the highly repetitive heterochromatic regions. A reference genome that is incomplete or incorrect can cause reads to go unaligned or to align to the wrong place, causing researchers to miss potential genetic contributors to disease. Some scientists hypothesize that an incomplete reference genome may partially explain why we still don’t fully understand the genetic causes of most diseases, even those that clearly have a genetic basis, as they are very often passed down from parent to child.
In addition, it has become clear that certain regions of the human genome with high genomic diversity are also poorly served by the reference genome. These regions have elevated levels of structural variation—complex sequence differences involving long stretches of DNA—compared to more-conserved regions of the genome. They are so highly variable that no single sequence could adequately represent the entire human species. Many sequences are so divergent that alignment algorithms do not recognize them as belonging anywhere in the reference genome. In recent releases, the GRC has distributed a small number of divergent alternative sequences for targeted regions, but these are infrequently used, in part because genomic tools have not quite caught up.
Enter the scientists on a quest for ultimate genome completeness. The initial goal: to complete the full sequence, telomere-to-telomere, of every chromosome of a single human genome. The ultimate goal: to construct a reference genome that can capture genetic variation in all regions of the genome for all humans.
Leaps in technology propel genome sequencing
The Sanger sequencing that the original HGP primarily relied on was a time- and resource-intensive but extremely accurate way to determine the sequence of a DNA fragment up to about 1,000 bases long. In the two decades following the HGP, a handful of different sequencing modalities have been developed (see illustration), and the Telomere-to-Telomere (T2T) Consortium has capitalized on this new technology to help finally complete the sequence of a human genome of each and every one of our paired 23 chromosomes.
Two of these new modalities, short-read sequencing by synthesis and long-read nanopore sequencing, proved very important to the T2T Consortium’s early assembly efforts. Both are much higher throughput than the now outdated Sanger sequencing, but each has its own limitations. Long-read sequencing can sequence DNA fragments of more than 10,000 bases, simplifying genome assembly but has a relatively high error rate. Short-read sequencing has a very low error rate but can only sequence 100 to 500 bases at a time. Using short reads, the 3-billion-base-pair human genome puzzle built from, say, 150-base-pair-long sequences means researchers are working with a 20-million-piece puzzle. And if alignment with the reference genome is putting together a puzzle with the help of the photo on the box, assembling genomes de novo is trying to do a puzzle bought at a garage sale that didn’t come with the box and may not even contain all of the pieces.
Some regions in the genome are highly repetitive, and when these repeats are longer than the fragments being sequenced, they are all but impossible to assemble. Imagine if that solid blue sky were made up of thousands of tiny pieces. Alternatively, long-read sequencing results in a puzzle with many fewer pieces—perhaps the sky now fits on a single piece—but the error rate of long-reads means that neighboring pieces may not actually fit together. Sometimes, researchers use combinations of both long- and short-read sequencing technologies to solve the assembly puzzle, with long reads providing a rough idea of the sequences in each region of the genome and short reads smoothing out the errors.
The T2T Consortium started out with the goal of assembling the X chromosome, the sex chromosome that appears in one copy in males and two in females. Using long-read sequencing, the researchers first developed a series of scaffolds—long stretches of sequence that they knew were probably error-prone. With the help of short-read sequencing, the researchers corrected the errors within each of those longer sequences. They then used optical mapping, which can determine the genomic distance between short DNA sequence motifs on the same chromosome, to figure out where each large piece of the puzzle belonged. It was a long, iterative process involving numerous sequencing technologies, manual curation, and repeated validation of the assembly. But in 2019, the T2T Consortium released the first-ever fully sequenced human chromosome, filling in 29 gaps and 1,147,861 bases of sequence.
Before the T2T Consortium followed the same painstaking process to complete the rest of the chromosomes, sequencing technology made another leap. Pacific Biosciences (PacBio) had been refining their single-molecule real-time (SMRT) sequencing devices, which use an engineered polymerase enzyme to add complementary fluorescently tagged nucleotides to a template strand of DNA. It can sequence fragments of more than 100 kb. When the company then developed a protocol to circularize a strand of DNA and allow the polymerase to run through a single sequence dozens of times to greatly improve accuracy, that took the cake. The T2T Consortium abandoned the pipeline it had used for the X chromosome assembly and relied almost entirely on this new, high-fidelity (HiFi) long-read sequencing technology.
The T2T Consortium assembled the rest of the chromosomes using almost exclusively HiFi sequencing data with the help of ultra-long-read nanopore sequencing (with reads close to a full megabase) to stitch together a few tricky contigs that contained highly repetitive sequences. Most previous assembly algorithms worked with either short reads or error-prone long reads, and they usually gave up on assembling those highly repetitive regions. Thus, the T2T Consortium had to invent new assembly algorithms that could work with the new type of data. Their innovation paid off, and less than two years after publishing the sequence of the X chromosome, they released their full genome assembly, filling in the 200 million base pairs of gaps in the autosomes and X chromosome of the human reference genome. In total, the new assembly contains 3,054,815,472 bases spread out over 23 chromosomes, plus 16,569 bases of mitochondrial DNA.
While groundbreaking, the T2T assembly is not perfect; there are still a handful of regions that may contain small sequencing errors. Even more importantly, the T2T assembly is the sequence of only a single genome—and a uniformly homozygous one at that. The T2T sequenced the genome from CHM13, a cell line derived from an abnormally fertilized egg that has two identical copies of each chromosome instead of the maternal and paternal copies of each chromosome that somatic human cells have. Thanks to CHM13, the T2T did not have to worry about resolving the subtly different sequences on each chromosomal copy, but their assembly pipeline cannot yet fully assemble genomes from somatic cells. (This also means that CHM13 has no Y chromosome, though the T2T has released a preliminary assembly of a Y chromosome from another cell line.) Moreover, while the T2T assembly fixes one aspect of incompleteness of the human genome, it does not address the other glaring issue plaguing the human reference genome: representation of genetic diversity.
The Evolution of SequencingThere have been numerous sequence modalities developed in the last quarter century, but major advances include Sanger sequencing, sequencing by synthesis, nanopore long-read sequencing from Oxford Nanopore, and, most recently, high-fidelity single-molecule real-time sequencing from PacBio. These differ in the length of reads they generate, their efficiency, and accuracy, with technologies generally evolving to support faster, cheaper, and more-precise sequencing. | |
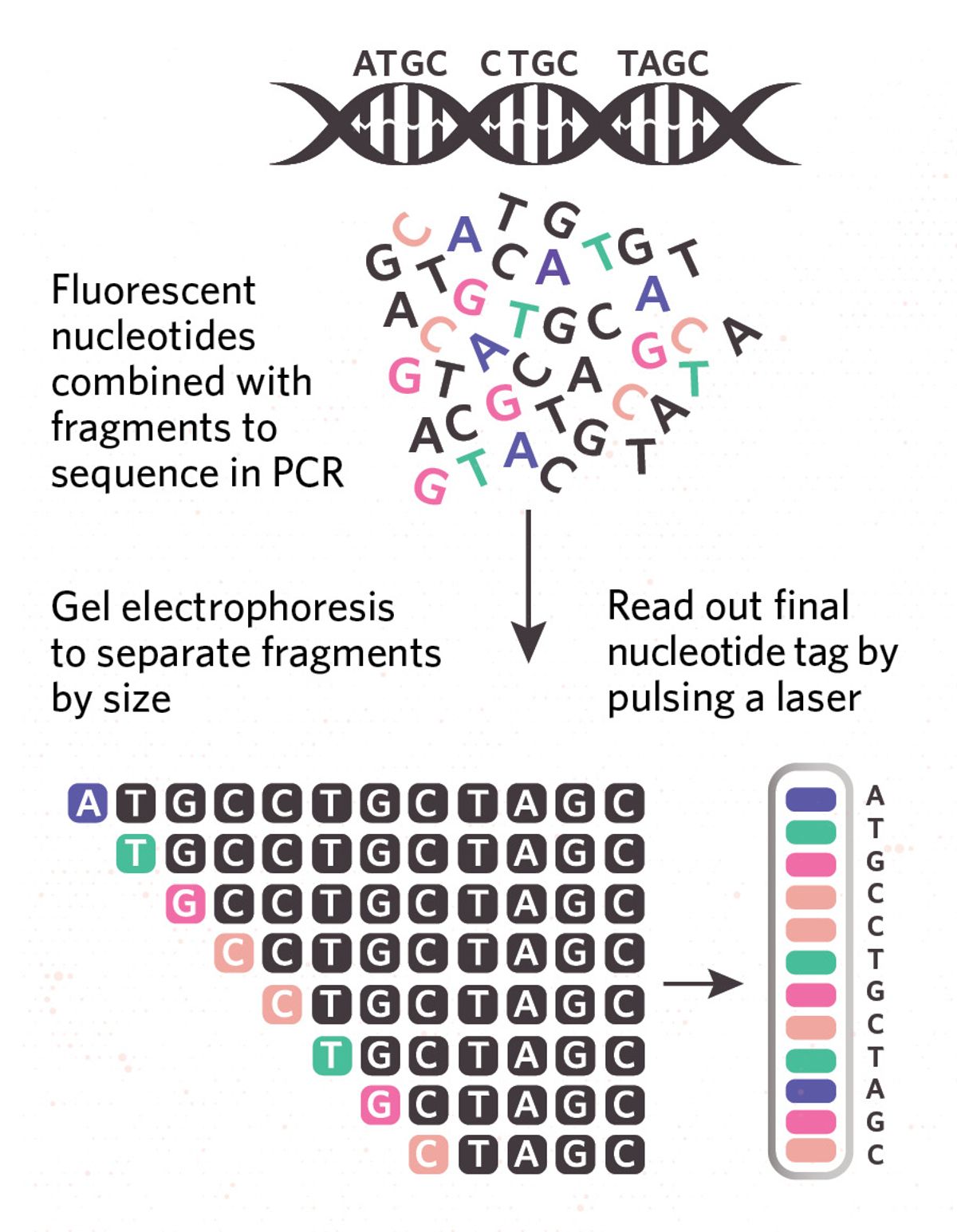 | SANGER SEQUENCINGThe first sequencing technology invented, and no longer used in modern projects, Sanger sequencing relies on tagging the ends of various sizes of DNA fragments with complementary fluorescent nucleotides. Fragments are then separated by size using gel electrophoresis and the final nucleotides’ fluorescence is read by a laser. The full sequence is inferred by piecing together the end nucleotides of the different-sized fragments. YEARS IN USE: 1980–2010 READ LENGTH: ~500–1,000 bases CONS: Low throughput, time intensive |
 | SEQUENCING BY SYNTHESISSequencing by synthesis (SBS) is the most commonly used type of sequencing today. It relies on synthesizing complementary DNA strands using fluorescently tagged nucleotides and capturing the output signal on a high-resolution camera. Hundreds of thousands of DNA fragments can be read at once, but SBS is limited to short lengths of DNA, making it challenging to assemble whole genomes de novo. YEARS IN USE: 2002–today READ LENGTH: ~100–500 bases CONS: Limited to short reads |
 modified from © istock.com, Dmitry Kovalchuk | NANOPORE SEQUENCINGOxford Nanopore devices pull DNA through a bioengineered pore to produce electrical current fluctuations that are then translated into a sequence. This approach generates long reads that can be used for de novo genome assembly or to identify larger structural variations that may not be possible with short reads, but it is less accurate than other sequencing technologies. YEARS IN USE: 2002–today READ LENGTH: ~10 kb–1 Mb CONS: Error-prone |
 | HIGH-FIDELITY SEQUENCINGOnly recently released by PacBio, high-fidelity (HiFi) single-molecule real-time (SMRT) sequencing relies on similar fluorescence strategies as SBS. Like nanopore sequencing, HiFi produces long reads that can be used for de novo genome assembly or to identify structural variants, but it achieves improved accuracy by circularizing a long DNA molecule so that it can be read dozens of times in a single run. YEARS IN USE: 2020–today READ LENGTH: ~10 kb CONS: Currently very expensive |
Genomics’ diversity problems
At the same time as the T2T Consortium was working to assemble the full human genome, genomicists had begun to reckon with the field’s lack of diversity. A large majority of human genomics data has come from participants in the US and Europe, with nearly 80 percent of GWAS participants being of European descent as of 2018.
There are several reasons for this sampling bias. Many of the first large sequencing centers were located in Europe or the United States. Additionally, the way the genomics research enterprise interfaces with potential study participants selects for people of certain socioethnic demographics: People with the time and resources to take part in large-scale genetic studies are more likely to come from wealthier regions of the world than from low- and middle-income countries. These factors also cause selection biases within the US and Europe, limiting participation in genomics research by people from underrepresented communities.
Exacerbating this shortcoming, researchers performing GWAS used to seek out populations with less overall genetic variation. Genomic data collection was expensive, and using such homogenous populations meant improved statistical power. Case in point: Iceland, one of the most homogenous populations in the world, is home to less than 0.1 percent of the world’s population but more than 10 percent of the world’s GWAS participants. On the other side of the spectrum, people of African ancestry, who have some of the most diverse genomes in the world, have historically been underrepresented in genomics studies.
Nearly 80 percent of GWAS participants were of European descent as of 2018.
Unfortunately, the bias in genomics studies has resulted in the identification of disease-associated loci primarily for European populations. We have an incomplete understanding of how or whether these same variations interact with different background genetic makeups to confer disease. We may also be missing opportunities to identify disease-associated variants that are rare in European populations but may be more common in people of other ancestries. Several groups have recognized the potential for this to lead to inequity in precision medicine and have launched projects to sequence the genomes of underrepresented or genetically diverse groups. Some researchers have argued that the wealth of genetic diversity in Africa provides an excellent opportunity to understand the relationship between human health and an enormous selection of different variations. Due to ancient human migration patterns, someone with African roots harbors a set of variants that are not only very different from those in European populations, but also very different from another person of African descent. Genomically speaking, sub-Saharan African and Northern African ancestries are about as different as sub-Saharan African and Northern European ancestries.
However, performing more GWAS on underrepresented groups does not entirely solve our diversity problem. GWAS is inherently limited to variants that can be described in terms of the reference genome. Many of the same groups whose variants have not been well-studied by GWAS also have large sections of genome that do not correspond well to any sequences on the reference genome. Using more than 900 individuals of African ancestry, one study identified nearly 300 million base pairs—nearly 10 percent of the length of the human genome—of new human DNA sequence not on the current reference genome. Variants in these regions are rendered effectively invisible using standard GWAS methods.
See “Sequences of African Genomes Highlights Long-Overlooked Diversity”
Such findings highlight an important fact about reference genomes. Even with a near-perfect assembly like the one produced by the T2T Consortium, there are still limitations to its ability to serve as a reference for an entire species. Some problems with reference genomes arise not from inaccuracies in the assembly, but rather from the use of a singular reference genome itself.
Toward a human pangenome
A single reference genome can only represent—at best—a single genome sequence from the population. The mismatch between a sampled genome and the reference genome leads to several technical problems collectively referred to as reference bias. Perhaps the most pernicious form of reference bias arises during the alignment step of sequencing experiments. If a sample’s genome sequence does not match the reference, the accuracy of read alignment degrades, with larger differences leading to greater errors. The result is that the analyses are systematically less accurate for genomes that are more different from the reference.
Reference bias has troubling implications. Namely, it can build ethnic biases into the foundations of genomics research methodologies. While the magnitude of reference bias is more difficult to assess than that of GWAS sampling bias, many experts in the field suspect that it is subtle but pervasive. And unlike GWAS sampling bias, reference bias cannot be solved simply by sampling a greater diversity of genomes. Substituting any other genome to serve as the reference sequence would result in just as much reference bias as the current reference genome. This was the core motivation behind the 2019 formation of the Human Pangenome Reference Consortium (HPRC), for which one of us (J.E.) has helped develop computational methods.
The HPRC has a broad purview with the end goal of producing a usable reference that includes most of the common DNA sequences in the human species, along with their context in the genome: a human pangenome. Luckily, the same technological advances that enabled the T2T human genome assembly are now breaking down the technological barriers that limited earlier pangenome projects.
Visualizing a PangenomeUnlike a linear reference genome, a graph genome allows a single region of the genome to take on a diverse set of sequences. For regions with high genetic diversity, a graph genome can better capture the many human DNA sequences that might exist.  Adapted from a graphic by Yohei Rosen |
Scientists are working on four major areas in parallel to achieve this goal. First, they must ascertain what human genetic variation exists. Because larger variation causes more-severe reference bias, there is a particular emphasis on identifying large, structural variation. Second, the HPRC aims to construct more-general reference structures that can express this variation. Third, the HPRC is building a set of tools that allow the reference pangenome to be used in practical settings. Finally, undergirding all this work, there is a concerted effort to consider the ethical, legal, and social implications of the research, from sampling all the way through to analysis and application.
Following the road paved by the T2T and smaller-scale efforts to catalog human structural variation, the HPRC is eschewing reference-based variant calling methodologies in favor of de novo genome assembly using long-read sequencing technology. To sample from the global human population, the HPRC cannot focus on a single homozygous cell line like the T2T Consortium did. Instead, it must perform the more challenging problem of assembling diploid genomes, in which the two copies of each chromosome differ. Because both copies are highly similar, it can be difficult to tease them apart. Thus far, the HPRC has tackled this challenge by focusing on mother-father-child trios, where a child’s two copies can be differentiated by comparing them to sequence data from their parents. The first phase of the HPRC genome assembly effort, completed in 2021, produced 45 diploid genome sequences, and there are plans to assemble 350 by the project’s completion in the next couple of years.
Representing a reference pangenome constructed with these data requires a data structure that is more expressive than a simple sequence. The most common alternatives include mathematical pangenome graphs. (See illustration.) These are formed by merging the shared sequence across many genomes. The resulting structure can be described by nodes that represent the sequence variations contained in a species’ genomes and edges that connect potential neighboring sequences together. Unlike conventional reference genomes, pangenome graphs can diverge and reconverge around variant sites, and they are expressive enough to capture genomic regions with complex structural variation. An individual’s genome can be described by traveling through the appropriate nodes of such a graph.
Even with a near-perfect assembly, there are still limitations to a genome’s ability to serve as a reference for an entire species.
Pangenome graph construction is currently a research frontier in bioinformatics. Data resources like the collection of assemblies produced by the HPRC are only just now becoming available, and the tools to utilize them are still being developed. The dust has not yet settled on which approaches will be most effective for which applications, but there is reason to hope that interindividual variation will soon be easy to characterize and reference bias will be largely a problem of the past. Already, early applications are showing improvements over the state of the art for some genomics analyses.
See “The Pangenome: Are Single Reference Genomes Dead?”
From where we stand now, the future of the human reference genome looks bright. Not only has the work of the T2T Consortium given humanity the first fully complete human genome sequence, the technology developed along the way has opened up new doors when it comes to sequencing and assembling genomes from diverse populations. Now, genomicists need to ensure that diversity and inclusion play a larger role in human genomics than it has in past decades, something that will be facilitated by the multinational collaborative culture put in place by the Human Genome Project and continued by the T2T, HPRC, and other global genomics consortia. With the participation of diverse institutions and participants, we hope to one day put forth a reference genome that represents not only the 3 billion bases of our mysterious man from Buffalo, but the full genomes of men and women from Brazil, Burkina Faso, Belarus, Bhutan, Botswana, Belize, Bangladesh, and beyond.
Brianna Chrisman is a final-year PhD student in the Bioengineering Department at Stanford University. Jordan Eizenga is a postdoctoral researcher in the University of California, Santa Cruz, Genomics Institute and a member of the Human Pangenome Reference Consortium.