Approximately 25 percent of proteins localize to membranes, where they act as receptors and signaling molecules in biological processes, including respiration, neurotransmission, and molecular transport. Because membrane proteins represent 40 percent of drug targets, understanding them is crucial for drug discovery. Yet, performing structure-function analyses on membrane proteins is challenging because in their native states they are embedded within the plasma membrane’s hydrophobic lipid bilayer. Because of this, membrane proteins are inherently unstable in aqueous solution, requiring distinct strategies for their solubilization, purification, and maintenance.1 Mass photometry is a bioanalytical characterization technology that can help researchers characterize membrane protein properties, such as oligomerization or other complex formations and assess the purity of samples containing membrane proteins.2
To preserve native membrane protein properties and structures, researchers typically use membrane mimetics to recreate a lipid environment in aqueous solutions. Widely-used membrane mimetics for protein purification and structure-function studies include detergents, lipid emulsions, nanodiscs, and amphipols. Detergents are the most common solubilization and purification agents; however, they can affect protein structure and function, and pose challenges for downstream analysis. Additionally, selecting the best detergent to obtain large quantities of active, homogeneous, and stable protein requires laborious trial and error. Even after careful optimization, detergents do not necessarily resemble the native lipid bilayer, and researchers do not always obtain stably solubilized membrane proteins with intact functions. Amphipols, nanodiscs, and and other complex membrane mimetics are more stabilizing than detergents, but their use demands additional purification steps, increasing protocol complexity.1,3
Membrane protein purification using mimetics requires numerous steps and different protocols depending on the protein of interest. It is challenging for researchers to assess the quality of mimetics and identify which samples contain highly concentrated, stable membrane proteins. To overcome these difficulties, researchers working with membrane proteins would benefit from a method that can quickly and accurately characterize sample quality and composition.1,3
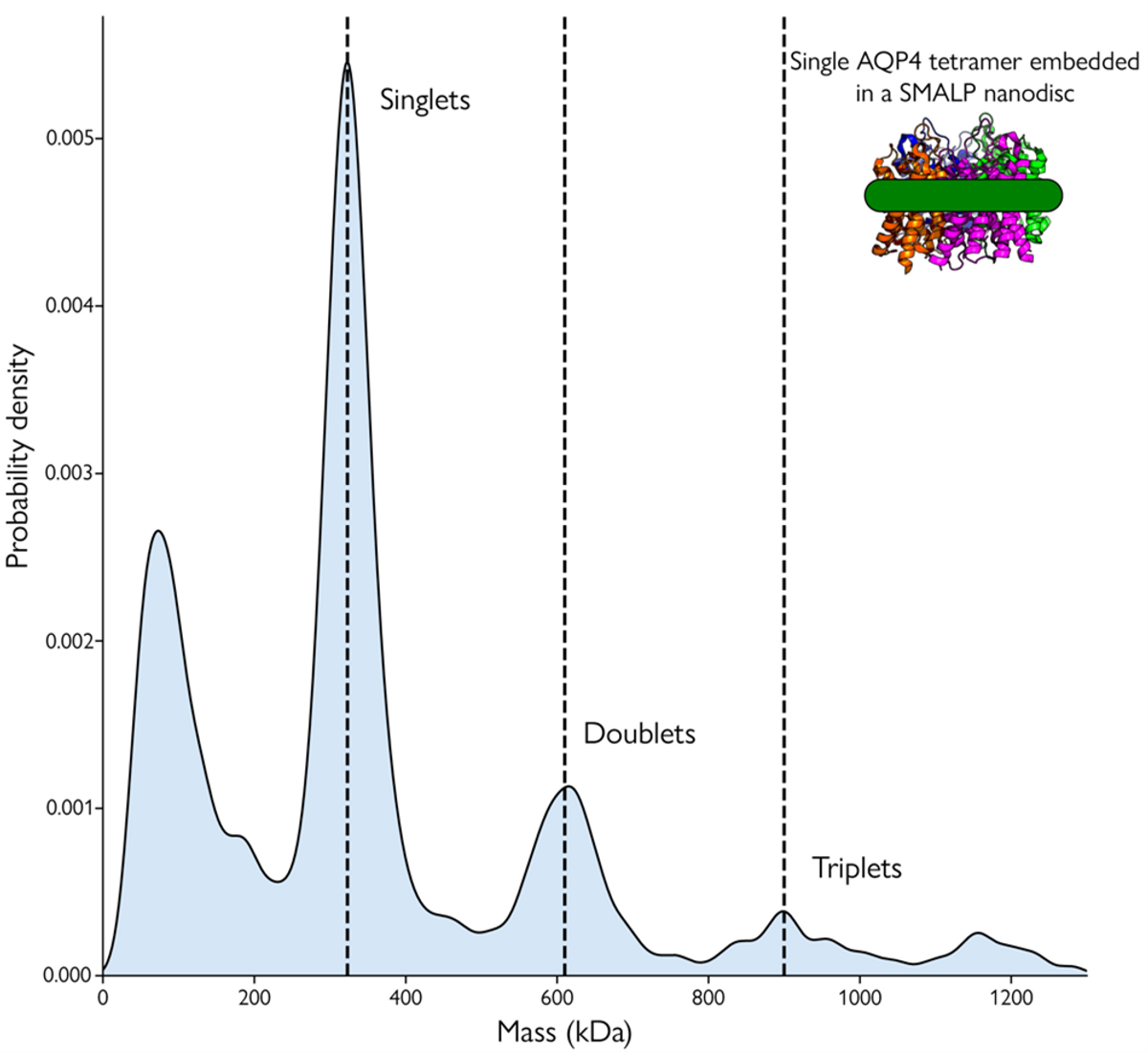
Mass photometry offers a powerful solution for the characterization of membrane proteins.2 The method uses a laser to measure the molecular mass of individual biomolecules in solution. A single molecule produces a measurable signal when illuminated by a laser, which is directly proportional to the molecule's mass. Mass photometry provides the mass distribution of membrane proteins and other sample components at the single-molecule level within a few minutes using very little sample. By quantifying the mass distribution of biomolecules in their samples, researchers gain valuable insights into sample purity, heterogeneity, and protein stoichiometry.4,5
Compared to other protein characterization techniques, such as SDS-PAGE and HPLC, mass photometry offers several unique advantages. It measures proteins’ true molecular mass instead of relying on surrogate measures to predict molecular weights. Moreover, mass photometry works with a variety of membrane proteins and mimetics including detergents, nanodiscs, styrene maleic acid copolymers form self-assembling lipid-protein particles (SMALPs), and amphipols, making it compatible with extant membrane protein purification workflows. Using this method, researchers can directly analyze samples without labeling or other modifications necessary for traditional methods, which can interfere with a membrane protein's structure, function, and activity.4-6
Using mass photometry to count and measure the mass of the single membrane protein in solution allows researchers to gain valuable information on the purity and stability of their samples. Information on aggregation allows them to ensure that they are working with solubilized samples. Oligomerization analyses help researchers identify and compare membrane protein states under different experimental conditions. Researchers also investigate biomolecular interactions involving membrane proteins using mass photometry. 5,6
References
- A. Pandey et al., “Current strategies for protein production and purification enabling membrane protein structural biology,” Biochem Cell Biol, 6:507-27, 2016.
- “How to study membrane proteins with mass photometry,” https://www.refeyn.com/post/how-to-study-membrane-proteins-with-mass-photometry#viewer-d10g2, accessed on December 27, 2022.
- E.P. Carpenter et al., “Overcoming the challenges of membrane protein crystallography,” Curr Opin Struct Biol, 5:581-86, 2008.
- “How does mass photometry work?,” https://www.refeyn.com/post/how-does-mass-photometry-work, accessed on December 27, 2022.
- A. Olerinyova et al., “Mass photometry of membrane proteins,” Chem, 7(1):224-36, 2021.
- Mass Photometry with detergents, https://www.refeyn.com/mass-photometry-with-detergents, accessed on December 27, 2022.
-1.jpg)