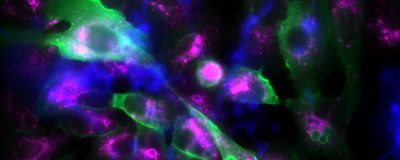
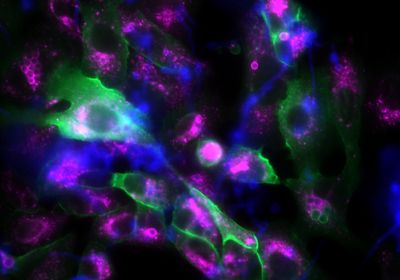
ABOVE: This fluorescence microscopy image shows epithelial cells (large, irregular structures) and fungal spores (small, spherical structures). If the fungal spores are surrounded by p11, they appear green. The violet fluorescence color, on the other hand, marks a protein in matured phagosomes. Leijie Jia/Leibniz-HKI
Fungal pathogens have emerged as a significant threat to human health as they become more prevalent and resistant to the handful of antifungal treatments available today.1 One of the toughest fungi to beat is Aspergillus fumigatus, a widespread mold that can cause life-threatening pulmonary infections in people with weakened immune systems.
A. fumigatus dodges the host immune system by sneaking into cells in membrane-enclosed vesicles known as phagosomes. These pathogen-containing vesicles are usually headed for degradation as the phagosomes mature and become more acidified. For A. fumigatus, though, intracellular extermination is not a done deal.
Scientists knew that the mold could interfere with the process of phagosome maturation; however, there was limited knowledge about the molecules used to set the fungal escape plan in motion. In a recent study, researchers discovered that A. fumigatus spores redirect fungus-containing phagosomes from a degradative to a non-degradative pathway by hijacking a human protein in those vesicles. They also found that transplant patients with a particular mutation in the gene that encodes the protein were less likely to develop invasive aspergillosis, a severe fungal infection with a high mortality rate among immunocompromised patients. The findings were published in the journal Cell Host & Microbe.2
Axel Brakhage, the scientific director of the Leibniz Institute for Natural Product Research and Infection Biology and study coauthor, has been interested in understanding how this fungus evades degradation inside the cell. His group previously found that a pigment present on the surface of A. fumigatus prevents phagosome maturation in immune cells.3 But they were surprised to find that some spore mutants lacking the pigment could still avoid phagocytic destruction, suggesting that the mold adopts an alternative intracellular escape strategy.
See also “Fungal Pathogens Flourish in the Pandemic’s Shadow”
To uncover A. fumigatus’s escape mechanism, the team assessed how the fungal spore proteins interacted with human lung epithelial cells, the body’s first line of defense against the mold. They showed that A. fumigatus spores use a surface protein, HscA, to bind to epithelial cells and that HscA was key to keeping the phagosomes in an immature state—less acidified vesicles that are not headed for degradation.
They next showed that HscA targets a small human protein found in phagosomes, which the researchers named p11. By interacting with p11, A. fumigatus spores alter the molecular marks on the phagosomes, excluding a mark for vesicle destruction and recruiting tags for phagosome recycling. In turn, this leads the vesicle to be released into the extracellular space, transferred to an adjacent cell, or to stay inside the cells.
For Scott Filler, a researcher at the University of California, Los Angeles, who was not involved in the study, the identification of HscA is a key finding. “This protein on the Aspergillus surface…basically stalls phagosome maturation,” he said.
Although epithelial lung cells are an important barrier against A. fumigatus, they are not specialized in killing pathogens as immune cells are. To investigate if the fungus would pull off its escape strategy in immune cells, the team repeated the experiments in macrophages and neutrophils. “We found basically the same findings; it is a general mechanism,” said Brakhage.
See also “Fungal DNA, Cells Found in Human Tumors”
To assess whether the in vitro findings were also clinically relevant, the researchers screened a group of stem cell transplant recipients and their corresponding donors for single nucleotide polymorphisms (SNP) in the p11 gene and evaluated their risk for developing invasive pulmonary aspergillosis (IPA). They found that a specific SNP located in a noncoding region of the p11 gene associated with a decreased risk for IPA. The fact that the researchers looked at patients to substantiate their in vitro findings makes the study stand out, said Filler. “It is very nice to go all the way from an in vitro model into basically human patients.”
The findings not only uncover the different components used by A. fumigatus to dodge intracellular destruction in the host’s cells, but they also provide clinical evidence that may help identify patients at greater risk for fungal infections who would benefit the most from antifungal treatments.
References
- www.who.int/news/item/25-10-2022-who-releases-first-ever-list-of-health-threatening-fungi
- Jia LJ, Rafiq M, Radosa L, et al. Aspergillus fumigatus hijacks human p11 to redirect fungal-containing phagosomes to non-degradative pathway. Cell Host Microbe. 2023; 31(3): 373-388.e10. doi:10.1016/j.chom.2023.02.002
- Thywißen A, Heinekamp T, Dahse HM, et al. Conidial dihydroxynaphthalene melanin of the human pathogenic fungus Aspergillus fumigatus interferes with the host endocytosis pathway. Front Microbiol. 2011; 2:96. Published 2011 May 3. doi:10.3389/fmicb.2011.00096