ABOVE: modified from © istock.com, BlackJack3D, Tetiana Lazunova
Life science has a habit of overturning long-standing ideas about the causes of human disease. Xin Lu, a cancer biologist at University of Notre Dame, came to that realization a few years ago when a family of five visited his institution to learn more about their condition. The father had been diagnosed with von Hippel-Lindau disease (VHL), a rare, inherited condition that causes slow-growing, benign tumors that typically occur in the central nervous system. Most of the man’s tumors could be surgically removed, but he had passed on the disease to two of his three children—twin girls who had developed inoperable tumors in their retinas at age seven.
Lu was familiar with the kinds of mutations that cause VHL. They typically involve nucleotide changes that alter the amino acid sequence of the VHL protein, rendering it dysfunctional, although sometimes the relevant gene is deleted entirely. The mutations usually don’t have an effect until the second gene copy from the unaffected parent is lost or silenced in certain tissues. The consequence is a scarcity of the VHL protein—which plays an important role in preventing uncontrolled cell proliferation and dysregulated blood vessel growth—leading to tumor formation.
However, when Lu looked at genomic sequencing data from the father and the twin girls, he was startled to find no protein-altering mutations in the VHL gene. There was only a single mutation on the one allele, but it seemed insignificant; it switched an adenine nucleotide into a guanine, changing the sequence “CCA” into “CCG.” As those two sequences code for the same amino acid—proline—the mutation shouldn’t make any difference, Lu reasoned. “We were puzzled for a while,” he recalled.
“Synonymous” mutations like these—ones that don’t alter the amino acid sequence—have often been dismissed as silent—mere genomic oddities without any biological effects. But through a series of analyses on cell lines isolated from skin samples from each family member, Lu learned that the VHL mutation nevertheless disrupted gene expression. It turned out that the mutation affected gene splicing. The proteins that carry out splicing bind to specific nucleotide sequences in order to excise the correct sequences. By muddling one of those splicing sites, the mutation caused the splicing machinery to miss a chunk of protein-coding RNA, creating shortened VHL transcripts. Consequently, the father and twin girls had no full-length VHL protein in particular tissues in which their healthy copy was lost—only a shorter, less stable version, Lu and his colleagues reported in 2020 in BMC Medical Genetics. “I didn’t know [that] these synonymous mutations could cause disease at all before I started this [study],” he said.
It’s very different mechanisms [via which] these mutations act.
—Kim De Keersmaecker, Katholieke Universiteit Leuven
In fact, the paper is part of a growing body of evidence that synonymous mutations aren’t always innocent. They’re often under selective pressure, can disrupt the process of protein-making in several ways, and have been implicated in conditions such as cystic fibrosis and autism, as well as cancer, where scientists have found that such mutations can disable key tumor suppressor genes like VHL. Still, their role in cancer isn’t well understood. They’re largely ignored in sequencing studies and are discarded from some databases of cancer mutations, with many researchers remaining focused primarily on nonsynonymous mutations, which likely have a greater overall impact in the cancer genome.
Some scientists are pushing to change this attitude, following estimates that certain synonymous alterations to DNA could account for 5–8 percent of all cancer-driving mutations. These researchers are now intensifying efforts to decipher the ways in which such mutations can trigger cancer and understand the scale of their impact across the genome through systematic studies. Such investigations could not only identify the disease-driving mechanisms for a portion of patients, but perhaps also open up new therapeutic avenues.
“I’m not advocating that every synonymous mutation does something. That’s not the case,” said Sven Diederichs, a molecular biologist at the German Cancer Research Center and the University of Freiburg. But “I would say there are definitely some that do something, and these are worth being investigated.”
How silent mutations make noise
Work such as Lu’s shows that synonymous codons aren’t entirely interchangeable. The precise codon can influence the binding of expression-regulating proteins and RNA to regulatory sites on DNA (which can also occur in coding regions) and affect the interactions of splicing machinery with pre-mRNA. For instance, some synonymous mutations encourage tumor growth by disrupting binding sequences for proteins and RNAs that usually repress gene expression. In the case of splicing disruptions—the best-studied and likely most consequential impact of synonymous mutations—the aberrations can not only abolish tumor suppressors through exon skipping but can also boost expression of cancer-driving genes, for instance by creating new sequences that promote splicing and lead to faster processing of mRNA.
Different synonymous codons also affect binding sites on the mRNA for proteins or microRNAs and how the mRNA folds into three-dimensional bends and loops, which in turn influence how long it lasts in the cytoplasm and how much of it is available to ribosomes. “The more folded it is, the more stable it is on average, and the more stable it is, the more times it will get translated,” explained bioinformatician Yana Bromberg of Emory University, whose research includes developing computer models for predicting the effects of synonymous mutations.
Codons also influence translation efficiency. Different species—and even different tissues and genes within an organism—preferentially use certain synonymous codons to encode particular amino acids to optimize their specific translation machinery. A rarely used synonymous codon in a given species will take longer to translate than a commonly used one, for instance, because the corresponding tRNAs that shuttle the amino acid in question during translation are less abundant. Slow translation can even alter how the protein folds as it exits the ribosome. Biochemists trying to express human genes in bacteria often account for this by tweaking gene sequences to match bacteria’s preferred codons to achieve optimal levels of protein, noted molecular biologist Olivia Rissland of the University of Colorado School of Medicine. In human cells, synonymous mutations have been observed to slow down translation by replacing commonly used codons with rare ones, or accelerating translation by switching rare ones for common ones—altering the structure of the final protein or its concentrations. Ultimately, Rissland said, “codon usage affects almost every aspect of gene expression.”

The upshot is that synonymous mutations can affect gene expression in a variety of ways that drive tumor growth. Interestingly, synonymous mutations don’t always act on their own—something that thoracic oncologist and cancer researcher Pasi Jänne of the Dana-Faber Cancer Institute found during a recent study that employed CRISPR to engineer various mutations into human cancer cell lines and examined those cells’ responses to cancer drugs. The team noticed that cell populations carrying a particular nonsynonymous mutation in the oncogene KRAS would only survive under drug exposure if they also carried a synonymous mutation. Further investigation revealed that on its own, the nonsynonymous mutation creates a new splice site that effectively removes a protein-coding sequence from the mRNA, abolishing the protein. But the synonymous mutation disrupts that misshapen splice site, causing the cancer-driving protein to be expressed after all. “We were surprised on multiple aspects of it,” Jänne said. “We’ll continue to pay more attention to these kinds of mutations if we ever stumble upon another one.”
Many discoveries of synonymous mutations having deleterious effects in cancer have been serendipitous, Rissland observed. What are missing are systematic, genome-wide studies to untangle the scale of these genetic alterations’ impact in cancer. The challenge there is sifting out the synonymous mutations that have an effect from the truly innocent ones—a much harder task than for nonsynonymous mutations where there are robust tools to predict their effect, noted cancer researcher Kim De Keersmaecker of the Katholieke Universiteit Leuven. With synonymous mutations, “you really need a completely different mindset,” she said. “It’s very different mechanisms [via which] these mutations act.”
NOT SO SILENT AFTER ALLAlthough scientists often assume that synonymous mutations don’t cause any biological effects because they don’t alter the amino acid sequence, recent research shows that they can influence transcription and translation in a variety of ways. | |
 MODIFIED FROM © ISTOCK.COM, TTSZ | |
 | ALTERING REGULATORY BINDING SITESAlthough a synonymous mutation may not alter the amino acid sequence of a protein, the change may still affect the binding of regulatory proteins that require specific sequences to interact with DNA. For instance, one not yet published study describes two synonymous mutations in the B-cell lymphoma (BCL2) gene in some lymphoma patients that appeared to prevent the binding of musculin, a transcription factor that represses expression of the gene. |
 MODIFIED FROM © ISTOCK.COM, TTSZ | |
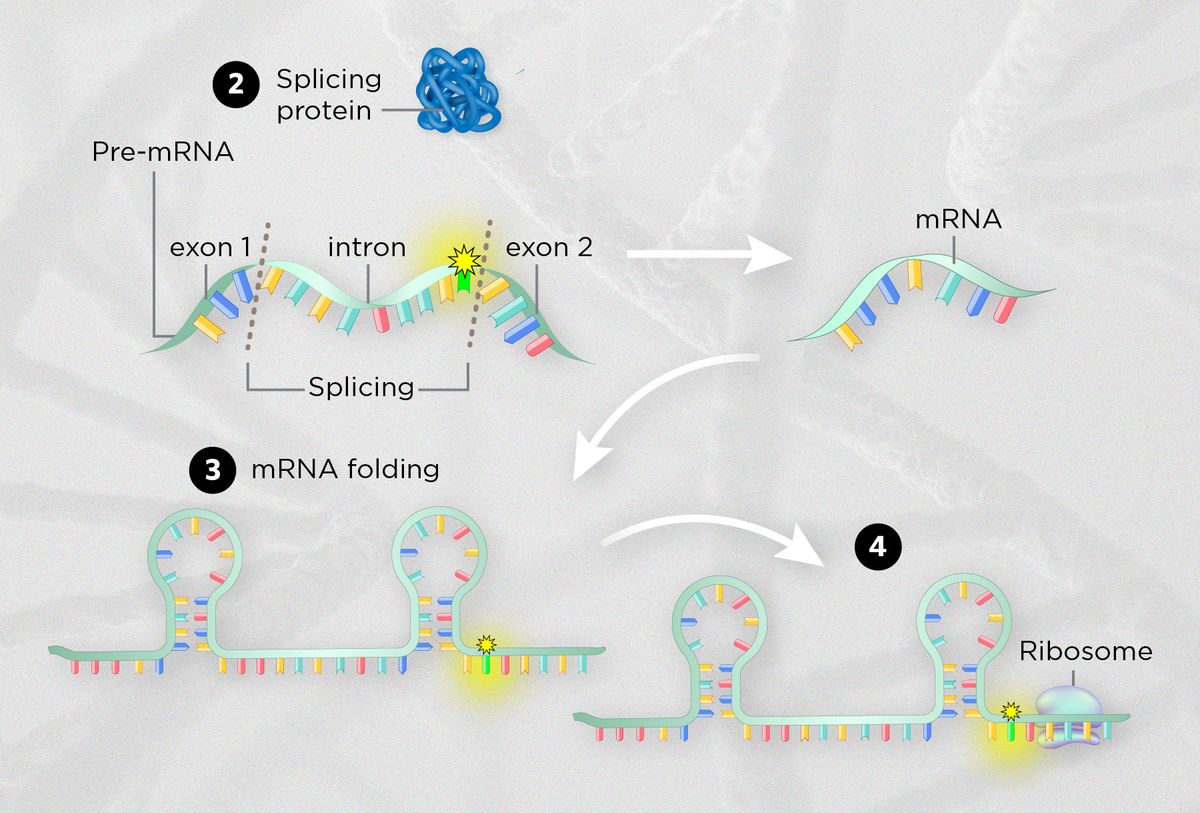
 | DISRUPTING SPLICING PATTERNSSynonymous mutations can remove binding sites for splicing proteins or create new ones, or change the rate of splicing overall. For instance, one recent study reported a synonymous mutation in the tumor suppressor gene BAP1 in a patient with a severe form of kidney cancer. The mutation likely causes the splicing machinery to miss a key piece of protein-coding DNA, effectively abolishing the protein that would normally help repress tumor growth. |
 | WARPING mRNA STRUCTUREChemical interactions between nucleotide bases cause mRNA to fold into three-dimensional structures that determine its overall stability. Synonymous mutations can affect binding sites on the mRNA for proteins and microRNAs, and also disrupt splicing or translation by affecting how the mRNA folds into three-dimensional structures. One 2003 study suggested that a synonymous mutation in the gene encoding the human dopamine receptor D2 makes the mRNA fold into a less stable conformation, causing less of it to be translated into protein. |
 | EFFECTS ON TRANSLATIONSynonymous mutations can also influence the speed of translation because the precise codon used determines how quickly it’s translated by ribosomes. For instance, one 2016 study reported that a handful of synonymous mutations in the KRAS oncogene changed a rare glycine-encoding codon into a commonly used one, potentially accelerating translation. Mutation-carrying cell lines had much higher protein levels than wild type cells and proliferated more quickly. |
Scanning the genome
Diederichs and his colleagues made some headway on that goal in a 2019 study in which they examined a database of nearly 3 million mutations identified from more than 18,000 tissue samples from 88 tumor types. From this, they assembled their own database comprising more than 650,000 synonymous mutations. Like nonsynonymous mutations, the synonymous ones tended to cluster in cancer-related genes—an indication that at least some of them may also be selectively advantageous for cancer cells. The team compiled numerous metrics to gauge whether a given mutation could have a cancer-driving effect, such as disrupting known splicing sites, and counted how often each mutation recurred across samples. They also used previously developed algorithms to estimate the probability of a mutation changing the structure of a gene’s mRNA transcript.
Based on this analysis, the team picked out a handful of synonymous mutations in the KRAS oncogene to study in further detail. One by one, they expressed the mutation-studded genes in human cell lines using plasmids. As expected, some mutations led to upregulation of levels of KRAS protein, which could hypothetically link to tumor formation. For one mutation, algorithms had predicted that the mRNA would fold differently, which could alter the gene’s expression. The team used a specialized technique to determine the actual structure of the mRNA by examining its chemical interactions with nucleotide-containing solutions. Sure enough, “we found exactly the same pattern that was predicted,” Diederichs said. To his knowledge, this is the first evidence that a synonymous mutation could change a gene’s mRNA structure in cancer.
Diederichs said that the field needs many more experimental studies like these to validate predictions of the effects of synonymous mutations. He acknowledged, however, that the technique he used to do so—expressing mutated genes in plasmids—isn’t ideal; for one, it introduces genetic material in an already spliced state, making it inadequate to assess the effects of mutations on splicing. It would be more useful to systematically introduce synonymous mutations into cells’ nuclear genomes, although doing that kind of work at scale would be labor intensive and technically challenging. “What we would really need is an analysis where you . . . have an experimental validation of like 10,000 synonymous mutations and see how many of them actually do something,” he said.
Codon usage affects almost every aspect of gene expression.
—Olivia Rissland, University of Colorado School of Medicine
Meanwhile, some scientists are focusing on finding new ways to predict the effects of synonymous mutations on gene expression, including possible impacts on protein translation—something that Rissland said can be trickier and more expensive to assess than effects on splicing, for instance. Computational and synthetic biologist Tamir Tuller of Tel Aviv University is building computational models that predict effects on translation initiation and speed, among models for other steps of gene expression, although he stressed that the subsequent effects on protein folding remains challenging. Molecular geneticist Helena Persson at Lund University, meanwhile, is incorporating new metrics to pick out possible translation-related effects. Using data from a cohort of roughly 3,400 breast cancer patients in Sweden, she’s developing methods to identify inherited silent mutations in known oncogenes that correlate with a person’s risk of developing familial breast cancer. Among other metrics, she’s quantifying how often mutations change commonly used codons into rarely used ones and vice versa; if a mutation has created a rare codon, for instance, it might slow translation and affect protein folding.
She and her colleagues have already begun to test the quality of their predictions on the gene encoding the estrogen receptor, a transcription factor implicated in breast cancer. They discovered one synonymous mutation that decreases the activity of the receptor under certain conditions by transforming a common codon into a rare one. Persson suspects that the mutation might slow down translation at a site where translation speed is key to getting the protein to fold correctly. Curiously, her preliminary results suggest that the mutation only has an effect where it co-occurs with certain other synonymous mutations, indicating that there could be important interactions between variants. “I think this is the most surprising thing I’ve learned about synonymous variants,” she added.
Studying how mutations interact with one another could be key to understanding not just the generation of tumors, but also their sensitivity and resistance to drugs, Diederichs said; such dynamics may help explain, for instance, why some patients with specific mutations don’t respond to drugs designed to target them. Persson said that her team is now planning to test combinations of synonymous variants rather than one variant at a time. “We have to actually take the whole transcript into consideration,” she said.
Practical applications
In addition to improving scientists’ general mechanistic understanding of cancer, Persson hopes that studying synonymous mutations will also bring benefits to patients—for example, by improving cancer screening among people who have a family history of breast cancer. Around 5–10 percent of breast cancer cases are hereditary, but scientists have only identified the disease-driving mutations for a small fraction of those. Synonymous mutations might “explain another couple of percent of these cases,” she said, and those mutations could then be included in preliminary cancer screening. The same approach may be relevant for other cancer types with a hereditary component, such as colon or ovarian cancer.
Tuller also sees a role for synonymous mutations in developing more general diagnostic and prognostic tools for cancer. In a 2021 study, he used a dataset of synonymous mutations identified in human cancer samples to train machine learning algorithms to predict the type of cancer in question as well as a patient’s likelihood of surviving 10 years after the initial diagnosis. When the team tested the algorithms’ predictive ability on a new subset of the data, Tuller was surprised to find that these algorithms performed just as well as algorithms that had been trained on non-synonymous mutations alone. “The results were stronger than . . . I expected,” Tuller said.
This predictive ability of synonymous mutations doesn’t necessarily point to a disease-driving effect; it could simply reflect general mutational patterns in the cancer genome—for instance ones produced by mutations in DNA replication enzymes. Either way, the study makes a compelling case for including silent mutations as biomarkers in tools used to classify tumors and identify appropriate therapies, Tuller said.
As therapeutic targets themselves, synonymous mutations are generally harder to treat than non-synonymous mutations, De Keersmaecker said. Most cancer therapies that target specific mutations are designed to inhibit pathogenic proteins that arise from mutated oncogenes, and most synonymous mutations probably don’t affect the structure of the final protein. However, even if a synonymous mutation doesn’t warp the protein structure, it perhaps could be tackled with the same treatments developed for nonsynonymous ones if the mutations have a similar effect on gene expression, Diederichs said. Consider a silent mutation that activates an oncogene—say, by removing a sequence in the gene that usually prevents its expression. “I would need to treat that patient exactly as if there was an activating [nonsynonymous] mutation in this gene,” Diederichs said; patients with such silent mutations could be candidates for drugs designed to inhibit the protein produced by that oncogene. In this way, studies of synonymous mutations could “broaden the spectrum of mutations that can actually be used for clinical decision making,” he said.
In the future, there may be ways of targeting other kinds of synonymous mutations such as those that cause changes to mRNA structure, De Keersmaecker said. Notably, in Jänne’s study, researchers had used an antisense oligonucleotide to block the effect of the synonymous mutation that caused the mutated KRAS gene to be expressed. Antisense oligonucleotides have been approved as therapies for other diseases such as to correct splicing in defective genes responsible for the degenerative disease Duchenne’s muscular dystrophy. But “whether that can be done therapeutically here is a different story,” Jänne said.
Cancer genomics has made major strides in recent decades in deciphering mechanisms and building therapies for the obvious low hanging fruit of nonsynonymous mutations. The study of synonymous mutations is part of a broader push to understand the more complex types of mutations that have been long overlooked. To Persson, the emerging evidence of their effect in cancer affirms that scientists should devote more attention to long ignored mechanisms of disease. “So many times before, people said, ‘it’s not worth looking at,’” Persson said. And then suddenly, “we find something that is really interesting and it opens up a new line of research.”